The eukaryotic translation initiation factor eIF4E elevates steady-state m7G capping of coding and noncoding transcripts
- PMID: 33055213
- PMCID: PMC7604501
- DOI: 10.1073/pnas.2002360117
The eukaryotic translation initiation factor eIF4E elevates steady-state m7G capping of coding and noncoding transcripts
Abstract
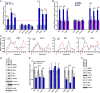
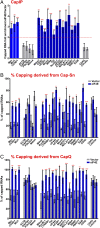
Methyl-7-guanosine (m7G) "capping" of coding and some noncoding RNAs is critical for their maturation and subsequent activity. Here, we discovered that eukaryotic translation initiation factor 4E (eIF4E), itself a cap-binding protein, drives the expression of the capping machinery and increased capping efficiency of ∼100 coding and noncoding RNAs. To quantify this, we developed enzymatic (cap quantification; CapQ) and quantitative cap immunoprecipitation (CapIP) methods. The CapQ method has the further advantage that it captures information about capping status independent of the type of 5' cap, i.e., it is not restricted to informing on m7G caps. These methodological advances led to unanticipated revelations: 1) Many RNA populations are inefficiently capped at steady state (∼30 to 50%), and eIF4E overexpression increased this to ∼60 to 100%, depending on the RNA; 2) eIF4E physically associates with noncoding RNAs in the nucleus; and 3) approximately half of eIF4E-capping targets identified are noncoding RNAs. eIF4E's association with noncoding RNAs strongly positions it to act beyond translation. Coding and noncoding capping targets have activities that influence survival, cell morphology, and cell-to-cell interaction. Given that RNA export and translation machineries typically utilize capped RNA substrates, capping regulation provides means to titrate the protein-coding capacity of the transcriptome and, for noncoding RNAs, to regulate their activities. We also discovered a cap sensitivity element (CapSE) which conferred eIF4E-dependent capping sensitivity. Finally, we observed elevated capping for specific RNAs in high-eIF4E leukemia specimens, supporting a role for cap dysregulation in malignancy. In all, levels of capping RNAs can be regulated by eIF4E.
Keywords: RNA capping; RNMT; eIF4E; methyl-7-guanosine cap.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
To cap it all off, again: dynamic capping and recapping of coding and non-coding RNAs to control transcript fate and biological activity.Cell Cycle. 2021 Jul;20(14):1347-1360. doi: 10.1080/15384101.2021.1930929. Epub 2021 Jul 9. Cell Cycle. 2021. PMID: 34241559 Free PMC article. Review.
-
Identification and Characterization of the Interaction Between the Methyl-7-Guanosine Cap Maturation Enzyme RNMT and the Cap-Binding Protein eIF4E.J Mol Biol. 2022 Mar 15;434(5):167451. doi: 10.1016/j.jmb.2022.167451. Epub 2022 Jan 10. J Mol Biol. 2022. PMID: 35026230 Free PMC article.
-
eIF4E orchestrates mRNA processing, RNA export and translation to modify specific protein production.Nucleus. 2024 Dec;15(1):2360196. doi: 10.1080/19491034.2024.2360196. Epub 2024 Jun 16. Nucleus. 2024. PMID: 38880976 Free PMC article. Review.
-
The nematode eukaryotic translation initiation factor 4E/G complex works with a trans-spliced leader stem-loop to enable efficient translation of trimethylguanosine-capped RNAs.Mol Cell Biol. 2010 Apr;30(8):1958-70. doi: 10.1128/MCB.01437-09. Epub 2010 Feb 12. Mol Cell Biol. 2010. PMID: 20154140 Free PMC article.
-
Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5' terminus.Biochemistry. 2004 May 11;43(18):5370-9. doi: 10.1021/bi030266t. Biochemistry. 2004. PMID: 15122903
Cited by
-
To cap it all off, again: dynamic capping and recapping of coding and non-coding RNAs to control transcript fate and biological activity.Cell Cycle. 2021 Jul;20(14):1347-1360. doi: 10.1080/15384101.2021.1930929. Epub 2021 Jul 9. Cell Cycle. 2021. PMID: 34241559 Free PMC article. Review.
-
M7G-related LncRNAs: A comprehensive analysis of the prognosis and immunity in glioma.Front Genet. 2022 Nov 16;13:961278. doi: 10.3389/fgene.2022.961278. eCollection 2022. Front Genet. 2022. PMID: 36468039 Free PMC article.
-
METTL3 alters capping enzyme expression and its activity on ribosomal proteins.bioRxiv [Preprint]. 2023 Nov 22:2023.11.22.568301. doi: 10.1101/2023.11.22.568301. bioRxiv. 2023. Update in: Sci Rep. 2024 Nov 12;14(1):27720. doi: 10.1038/s41598-024-78152-5. PMID: 38045284 Free PMC article. Updated. Preprint.
-
Nuclear RNA cap-chaperones eIF4E and NCBP2 govern distinct fates for 1000s of mRNAs uncovering an unexpected regulatory point in gene expression.bioRxiv [Preprint]. 2025 Jul 31:2025.07.25.666897. doi: 10.1101/2025.07.25.666897. bioRxiv. 2025. PMID: 40766477 Free PMC article. Preprint.
-
M7G-related tumor immunity: novel insights of RNA modification and potential therapeutic targets.Int J Biol Sci. 2024 Jan 27;20(4):1238-1255. doi: 10.7150/ijbs.90382. eCollection 2024. Int J Biol Sci. 2024. PMID: 38385078 Free PMC article. Review.
References
-
- Pillutla R. C., Shimamoto A., Furuichi Y., Shatkin A. J., Human mRNA capping enzyme (RNGTT) and cap methyltransferase (RNMT) map to 6q16 and 18p11.22-p11.23, respectively. Genomics 54, 351–353 (1998). - PubMed
-
- Fabrega C., Hausmann S., Shen V., Shuman S., Lima C. D., Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell 13, 77–89 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

