The Inhaled Steroid Ciclesonide Blocks SARS-CoV-2 RNA Replication by Targeting the Viral Replication-Transcription Complex in Cultured Cells
- PMID: 33055254
- PMCID: PMC7737752
- DOI: 10.1128/JVI.01648-20
The Inhaled Steroid Ciclesonide Blocks SARS-CoV-2 RNA Replication by Targeting the Viral Replication-Transcription Complex in Cultured Cells
Abstract
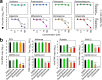
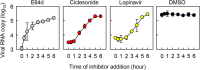
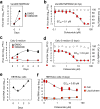
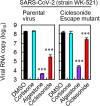
Here, we screened steroid compounds to obtain a drug expected to block host inflammatory responses and Middle East respiratory syndrome coronavirus (MERS-CoV) replication. Ciclesonide, an inhaled corticosteroid, suppressed the replication of MERS-CoV and other coronaviruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), in cultured cells. The 90% effective concentration (EC90) of ciclesonide for SARS-CoV-2 in differentiated human bronchial tracheal epithelial cells was 0.55 μM. Eight consecutive passages of 43 SARS-CoV-2 isolates in the presence of ciclesonide generated 15 resistant mutants harboring single amino acid substitutions in nonstructural protein 3 (nsp3) or nsp4. Of note, ciclesonide suppressed the replication of all these mutants by 90% or more, suggesting that these mutants cannot completely overcome ciclesonide blockade. Under a microscope, the viral RNA replication-transcription complex in cells, which is thought to be detectable using antibodies specific for nsp3 and double-stranded RNA, was observed to fall in the presence of ciclesonide in a concentration-dependent manner. These observations indicate that the suppressive effect of ciclesonide on viral replication is specific to coronaviruses, highlighting it as a candidate drug for the treatment of COVID-19 patients.IMPORTANCE The outbreak of SARS-CoV-2, the cause of COVID-19, is ongoing. New and effective antiviral agents that combat the disease are needed urgently. Here, we found that an inhaled corticosteroid, ciclesonide, suppresses the replication of coronaviruses, including betacoronaviruses (murine hepatitis virus type 2 [MHV-2], MERS-CoV, SARS-CoV, and SARS-CoV-2) and an alphacoronavirus (human coronavirus 229E [HCoV-229E]), in cultured cells. Ciclesonide is safe; indeed, it can be administered to infants at high concentrations. Thus, ciclesonide is expected to be a broad-spectrum antiviral drug that is effective against many members of the coronavirus family. It could be prescribed for the treatment of MERS and COVID-19.
Keywords: COVID-19; DMV; RTC; SARS-CoV-2; ciclesonide; coronavirus; steroid.
Copyright © 2020 American Society for Microbiology.
Figures




References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M-D, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group . 8 October 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med doi:10.1056/NEJMoa2007764. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

