The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections
- PMID: 33055260
- PMCID: PMC7565897
- DOI: 10.1128/mSphere.00909-20
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections
Abstract
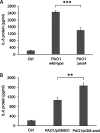
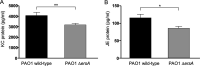
Bacterial small RNAs play a remarkable role in the regulation of functions involved in host-pathogen interaction. ErsA is a small RNA of Pseudomonas aeruginosa that contributes to the regulation of bacterial virulence traits such as biofilm formation and motility. Shown to take part in a regulatory circuit under the control of the envelope stress response sigma factor σ22, ErsA targets posttranscriptionally the key virulence-associated gene algC Moreover, ErsA contributes to biofilm development and motility through the posttranscriptional modulation of the transcription factor AmrZ. Intending to evaluate the regulatory relevance of ErsA in the pathogenesis of respiratory infections, we analyzed the impact of ErsA-mediated regulation on the virulence potential of P. aeruginosa and the stimulation of the inflammatory response during the infection of bronchial epithelial cells and a murine model. Furthermore, we assessed ErsA expression in a collection of P. aeruginosa clinical pulmonary isolates and investigated the link of ErsA with acquired antibiotic resistance by generating an ersA gene deletion mutant in a multidrug-resistant P. aeruginosa strain which has long been adapted in the airways of a cystic fibrosis (CF) patient. Our results show that the ErsA-mediated regulation is relevant for the P. aeruginosa pathogenicity during acute infection and contributes to the stimulation of the host inflammatory response. Besides, ErsA was able to be subjected to selective pressure for P. aeruginosa pathoadaptation and acquirement of resistance to antibiotics commonly used in clinical practice during chronic CF infections. Our findings establish the role of ErsA as an important regulatory element in the host-pathogen interaction.IMPORTANCEPseudomonas aeruginosa is one of the most critical multidrug-resistant opportunistic pathogens in humans, able to cause both lethal acute and chronic lung infections. Thorough knowledge of the regulatory mechanisms involved in the establishment and persistence of the airways infections by P. aeruginosa remains elusive. Emerging candidates as molecular regulators of pathogenesis in P. aeruginosa are small RNAs, which act posttranscriptionally as signal transducers of host cues. Known for being involved in the regulation of biofilm formation and responsive to envelope stress response, we show that the small RNA ErsA can play regulatory roles in acute infection, stimulation of host inflammatory response, and mechanisms of acquirement of antibiotic resistance and adaptation during the chronic lung infections of cystic fibrosis patients. Elucidating the complexity of the networks regulating host-pathogen interactions is crucial to identify novel targets for future therapeutic applications.
Keywords: ErsA; Pseudomonas aeruginosa; antibiotic resistance; clinical isolates; cystic fibrosis; mouse model of infection; opportunistic infections; pathogenicity; respiratory infections; small RNAs; virulence.
Copyright © 2020 Ferrara et al.
Figures



Similar articles
-
The Small RNA ErsA Impacts the Anaerobic Metabolism of Pseudomonas aeruginosa Through Post-Transcriptional Modulation of the Master Regulator Anr.Front Microbiol. 2021 Aug 20;12:691608. doi: 10.3389/fmicb.2021.691608. eCollection 2021. Front Microbiol. 2021. PMID: 34759894 Free PMC article.
-
Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts.PLoS One. 2012;7(4):e35648. doi: 10.1371/journal.pone.0035648. Epub 2012 Apr 25. PLoS One. 2012. PMID: 22558188 Free PMC article.
-
Positive signature-tagged mutagenesis in Pseudomonas aeruginosa: tracking patho-adaptive mutations promoting airways chronic infection.PLoS Pathog. 2011 Feb 3;7(2):e1001270. doi: 10.1371/journal.ppat.1001270. PLoS Pathog. 2011. PMID: 21304889 Free PMC article.
-
Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung.Curr Top Microbiol Immunol. 2013;358:91-118. doi: 10.1007/82_2011_199. Curr Top Microbiol Immunol. 2013. PMID: 22311171 Review.
-
Small Noncoding Regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia Complex.Int J Mol Sci. 2018 Nov 27;19(12):3759. doi: 10.3390/ijms19123759. Int J Mol Sci. 2018. PMID: 30486355 Free PMC article. Review.
Cited by
-
The function of small RNA in Pseudomonas aeruginosa.PeerJ. 2022 Jul 21;10:e13738. doi: 10.7717/peerj.13738. eCollection 2022. PeerJ. 2022. PMID: 35891650 Free PMC article.
-
Hfq-binding small RNA PqsS regulates Pseudomonas aeruginosa pqs quorum sensing system and virulence.NPJ Biofilms Microbiomes. 2024 Sep 11;10(1):82. doi: 10.1038/s41522-024-00550-4. NPJ Biofilms Microbiomes. 2024. PMID: 39261499 Free PMC article.
-
The role of RNA regulators, quorum sensing and c-di-GMP in bacterial biofilm formation.FEBS Open Bio. 2023 Jun;13(6):975-991. doi: 10.1002/2211-5463.13389. Epub 2022 Mar 13. FEBS Open Bio. 2023. PMID: 35234364 Free PMC article. Review.
-
Specific and Global RNA Regulators in Pseudomonas aeruginosa.Int J Mol Sci. 2021 Aug 11;22(16):8632. doi: 10.3390/ijms22168632. Int J Mol Sci. 2021. PMID: 34445336 Free PMC article. Review.
-
The Small RNA ErsA Impacts the Anaerobic Metabolism of Pseudomonas aeruginosa Through Post-Transcriptional Modulation of the Master Regulator Anr.Front Microbiol. 2021 Aug 20;12:691608. doi: 10.3389/fmicb.2021.691608. eCollection 2021. Front Microbiol. 2021. PMID: 34759894 Free PMC article.
References
-
- Wagner VE, Filiatrault MJ, Picardo KF, Iglewski BH. 2008. Pseudomonas aeruginosa virulence and pathogenesis issues, p 129–158. In Cornelis P (ed), Pseudomonas genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.

