Persistence of an intact HIV reservoir in phenotypically naive T cells
- PMID: 33055422
- PMCID: PMC7605525
- DOI: 10.1172/jci.insight.133157
Persistence of an intact HIV reservoir in phenotypically naive T cells
Abstract
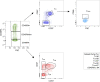
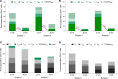
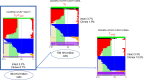
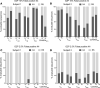
Despite the efficacy of antiretroviral therapy (ART), HIV persists in a latent form and remains a hurdle to eradication. CD4+ T lymphocytes harbor the majority of the HIV reservoir, but the role of individual subsets remains unclear. CD4+ T cells were sorted into central, transitional, effector memory, and naive T cells. We measured HIV DNA and performed proviral sequencing of more than 1900 proviruses in 2 subjects at 2 and 9 years after ART initiation to estimate the contribution of each subset to the reservoir. Although our study was limited to 2 subjects, we obtained comparable findings with publicly available sequences. While the HIV integration levels were lower in naive compared with memory T cells, naive cells were a major contributor to the intact proviral reservoir. Notably, proviral sequences isolated from naive cells appeared to be unique, while those retrieved from effector memory cells were mainly clonal. The number of clones increased as cells differentiated from a naive to an effector memory phenotype, suggesting naive cells repopulate the effector memory reservoir as previously shown for central memory cells. Naive T cells contribute substantially to the intact HIV reservoir and represent a significant hurdle for HIV eradication.
Keywords: AIDS/HIV; T cells.
Conflict of interest statement
Figures


Similar articles
-
Genetic Diversity, Compartmentalization, and Age of HIV Proviruses Persisting in CD4+ T Cell Subsets during Long-Term Combination Antiretroviral Therapy.J Virol. 2020 Feb 14;94(5):e01786-19. doi: 10.1128/JVI.01786-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31776273 Free PMC article.
-
Naive infection predicts reservoir diversity and is a formidable hurdle to HIV eradication.JCI Insight. 2021 Aug 23;6(16):e150794. doi: 10.1172/jci.insight.150794. JCI Insight. 2021. PMID: 34228640 Free PMC article.
-
Lack of concordance between residual viremia and viral variants driving de novo infection of CD4(+) T cells on ART.Retrovirology. 2016 Aug 2;13(1):51. doi: 10.1186/s12977-016-0282-9. Retrovirology. 2016. PMID: 27484989 Free PMC article.
-
Targeting the Latent Reservoir for HIV-1.Immunity. 2018 May 15;48(5):872-895. doi: 10.1016/j.immuni.2018.04.030. Immunity. 2018. PMID: 29768175 Free PMC article. Review.
-
Blocking Formation of the Stable HIV Reservoir: A New Perspective for HIV-1 Cure.Front Immunol. 2019 Aug 22;10:1966. doi: 10.3389/fimmu.2019.01966. eCollection 2019. Front Immunol. 2019. PMID: 31507594 Free PMC article. Review.
Cited by
-
Lentiviral Nef Proteins Differentially Govern the Establishment of Viral Latency.J Virol. 2022 Apr 13;96(7):e0220621. doi: 10.1128/jvi.02206-21. Epub 2022 Mar 10. J Virol. 2022. PMID: 35266804 Free PMC article.
-
Effects of COVID-19 mRNA vaccination on HIV viremia and reservoir size.AIDS. 2024 Jul 1;38(8):1120-1130. doi: 10.1097/QAD.0000000000003841. Epub 2024 Jan 22. AIDS. 2024. PMID: 38224350 Free PMC article.
-
The Clonal Expansion Dynamics of the HIV-1 Reservoir: Mechanisms of Integration Site-Dependent Proliferation and HIV-1 Persistence.Viruses. 2021 Sep 17;13(9):1858. doi: 10.3390/v13091858. Viruses. 2021. PMID: 34578439 Free PMC article. Review.
-
Prolonged non-suppressible viremia sustained by a clonally expanded, genomically defective provirus with an immune-evasive HIV protein expression profile.bioRxiv [Preprint]. 2025 May 16:2025.05.16.654538. doi: 10.1101/2025.05.16.654538. bioRxiv. 2025. PMID: 40462973 Free PMC article. Preprint.
-
Isotretinoin promotes elimination of translation-competent HIV latent reservoirs in CD4T cells.PLoS Pathog. 2024 Oct 14;20(10):e1012601. doi: 10.1371/journal.ppat.1012601. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39401241 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

