Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations
- PMID: 33056981
- PMCID: PMC7560655
- DOI: 10.1038/s41467-020-18987-4
Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations
Abstract
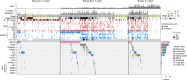
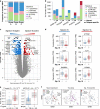
Neuroblastoma is a pediatric malignancy with heterogeneous clinical outcomes. To better understand neuroblastoma pathogenesis, here we analyze whole-genome, whole-exome and/or transcriptome data from 702 neuroblastoma samples. Forty percent of samples harbor at least one recurrent driver gene alteration and most aberrations, including MYCN, ATRX, and TERT alterations, differ in frequency by age. MYCN alterations occur at median 2.3 years of age, TERT at 3.8 years, and ATRX at 5.6 years. COSMIC mutational signature 18, previously associated with reactive oxygen species, is the most common cause of driver point mutations in neuroblastoma, including most ALK and Ras-activating variants. Signature 18 appears early and is continuous throughout disease evolution. Signature 18 is enriched in neuroblastomas with MYCN amplification, 17q gain, and increased expression of mitochondrial ribosome and electron transport-associated genes. Recurrent FGFR1 variants in six patients, and ALK N-terminal structural alterations in five samples, identify additional patients potentially amenable to precision therapy.
Conflict of interest statement
N.V.C. reports receiving commercial research grants from Y-mabs Therapeutics and Abpro-Labs Inc., holding ownership interest/equity in Y-Mabs Therapeutics Inc., holding ownership interest/equity in Abpro-Labs, and owning stock options in Eureka Therapeutics. N.V.C. is the inventor of issued and pending patents filed by Memorial Sloan-Kettering Cancer Center (MSK), including those licensed by MSK to Ymabs Therapeutics, Biotec Pharmacon, and Abpro-labs. Both MSK and N.V.C. have financial interest in Y-mabs. N.V.C. is an advisory board member for Abpro-Labs and Eureka Therapeutics. The remaining authors declare no competing interests.
Figures



References
-
- Simon T, Spitz R, Faldum A, Hero B, Berthold F. New definition of low-risk neuroblastoma using stage, age, and 1p and MYCN status. J. Pediatr. Hematol. Oncol. 2004;26:791–796. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

