Genetic alterations of SUGP1 mimic mutant-SF3B1 splice pattern in lung adenocarcinoma and other cancers
- PMID: 33057152
- PMCID: PMC7790757
- DOI: 10.1038/s41388-020-01507-5
Genetic alterations of SUGP1 mimic mutant-SF3B1 splice pattern in lung adenocarcinoma and other cancers
Abstract
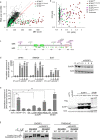
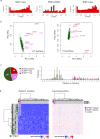
Genes involved in 3'-splice site recognition during mRNA splicing constitute an emerging class of oncogenes. SF3B1 is the most frequently mutated splicing factor in cancer, and SF3B1 mutants corrupt branchpoint recognition leading to usage of cryptic 3'-splice sites and subsequent aberrant junctions. For a comprehensive determination of alterations leading to this splicing pattern, we performed a pan-TCGA screening for SF3B1-specific aberrant acceptor usage. While the most of aberrant 3'-splice patterns were explained by SF3B1 mutations, we also detected nine SF3B1 wild-type tumors (including five lung adenocarcinomas). Genomic profile analysis of these tumors identified somatic mutations combined with loss-of-heterozygosity in the splicing factor SUGP1 in five of these cases. Modeling of SUGP1 loss and mutations in cell lines showed that both alterations induced mutant-SF3B1-like aberrant splicing. Our study provides definitive evidence that genetic alterations of SUGP1 genocopy SF3B1 mutations in lung adenocarcinoma and other cancers.
Conflict of interest statement
AH and MHS are inventors of a patent pending related to
Figures

References
-
- Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. 2019;9:1493–510. doi: 10.1158/2159-8290.CD-19-0399. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

