Fgf10-CRISPR mosaic mutants demonstrate the gene dose-related loss of the accessory lobe and decrease in the number of alveolar type 2 epithelial cells in mouse lung
- PMID: 33057360
- PMCID: PMC7561199
- DOI: 10.1371/journal.pone.0240333
Fgf10-CRISPR mosaic mutants demonstrate the gene dose-related loss of the accessory lobe and decrease in the number of alveolar type 2 epithelial cells in mouse lung
Abstract
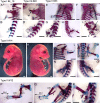
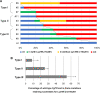
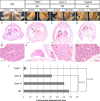
CRISPR/Cas9-mediated gene editing often generates founder generation (F0) mice that exhibit somatic mosaicism in the targeted gene(s). It has been known that Fibroblast growth factor 10 (Fgf10)-null mice exhibit limbless and lungless phenotypes, while intermediate limb phenotypes (variable defective limbs) are observed in the Fgf10-CRISPR F0 mice. However, how the lung phenotype in the Fgf10-mosaic mutants is related to the limb phenotype and genotype has not been investigated. In this study, we examined variable lung phenotypes in the Fgf10-targeted F0 mice to determine if the lung phenotype was correlated with percentage of functional Fgf10 genotypes. Firstly, according to a previous report, Fgf10-CRISPR F0 embryos on embryonic day 16.5 (E16.5) were classified into three types: type I, no limb; type II, limb defect; and type III, normal limbs. Cartilage and bone staining showed that limb truncations were observed in the girdle, (type I), stylopodial, or zeugopodial region (type II). Deep sequencing of the Fgf10-mutant genomes revealed that the mean proportion of codons that encode putative functional FGF10 was 8.3 ± 6.2% in type I, 25.3 ± 2.7% in type II, and 54.3 ± 9.5% in type III (mean ± standard error of the mean) mutants at E16.5. Histological studies showed that almost all lung lobes were absent in type I embryos. The accessory lung lobe was often absent in type II embryos with other lobes dysplastic. All lung lobes formed in type III embryos. The number of terminal tubules was significantly lower in type I and II embryos, but unchanged in type III embryos. To identify alveolar type 2 epithelial (AECII) cells, known to be reduced in the Fgf10-heterozygous mutant, immunostaining using anti-surfactant protein C (SPC) antibody was performed: In the E18.5 lungs, the number of AECII was correlated to the percentage of functional Fgf10 genotypes. These data suggest the Fgf10 gene dose-related loss of the accessory lobe and decrease in the number of alveolar type 2 epithelial cells in mouse lung. Since dysfunction of AECII cells has been implicated in the pathogenesis of parenchymal lung diseases, the Fgf10-CRISPR F0 mouse would present an ideal experimental system to explore it.
Conflict of interest statement
Supported by an academic grant from Pfizer Japan, Inc. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

