Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies
- PMID: 33057452
- PMCID: PMC7561147
- DOI: 10.1371/journal.pone.0240653
Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies
Abstract
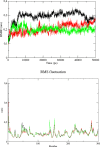
A novel coronavirus responsible of acute respiratory infection closely related to SARS-CoV has recently emerged. So far there is no consensus for drug treatment to stop the spread of the virus. Discovery of a drug that would limit the virus expansion is one of the biggest challenges faced by the humanity in the last decades. In this perspective, to test existing drugs as inhibitors of SARS-CoV-2 main protease is a good approach. Among natural phenolic compounds found in plants, fruit, and vegetables; flavonoids are the most abundant. Flavonoids, especially in their glycosylated forms, display a number of physiological activities, which makes them interesting to investigate as antiviral molecules. The flavonoids chemical structures were downloaded from PubChem and protease structure 6LU7 was from the Protein Data Bank site. Molecular docking study was performed using AutoDock Vina. Among the tested molecules Quercetin-3-O-rhamnoside showed the highest binding affinity (-9,7 kcal/mol). Docking studies showed that glycosylated flavonoids are good inhibitors for the SARS-CoV-2 protease and could be further investigated by in vitro and in vivo experiments for further validation. MD simulations were further performed to evaluate the dynamic behavior and stability of the protein in complex with the three best hits of docking experiments. Our results indicate that the rutin is a potential drug to inhibit the function of Chymotrypsin-like protease (3CL pro) of Coronavirus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Ahmad S, Abbasi HW, Shahid S, Gul S, Abbasi SW. Molecular Docking, Simulation and MM-PBSA Studies of Nigella Sativa Compounds: A Computational Quest to identify Potential Natural Antiviral for COVID-19 Treatment. Journal of Biomolecular Structure and Dynamics. 2020;(just-accepted):1–16. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

