The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET
- PMID: 33058760
- PMCID: PMC7557167
- DOI: 10.1016/j.str.2020.10.001
The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET
Abstract
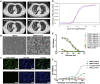
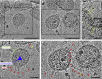
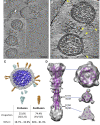
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) resulted from the outbreak of SARS-CoV-2 in December 2019. Currently, multiple efforts are being made to rapidly develop vaccines and treatments to fight COVID-19. Current vaccine candidates use inactivated SARS-CoV-2 viruses; therefore, it is important to understand the architecture of inactivated SARS-CoV-2. We have genetically and structurally characterized β-propiolactone-inactivated viruses from a propagated and purified clinical strain of SARS-CoV-2. We observed that the virus particles are roughly spherical or moderately pleiomorphic. Although a small fraction of prefusion spikes are found, most spikes appear nail shaped, thus resembling a postfusion state, where the S1 protein of the spike has disassociated from S2. Cryoelectron tomography and subtomogram averaging of these spikes yielded a density map that closely matches the overall structure of the SARS-CoV postfusion spike and its corresponding glycosylation site. Our findings have major implications for SARS-CoV-2 vaccine design, especially those using inactivated viruses.
Keywords: COVID-19; SARS-CoV-2; cryo-EM; cryo-ET; glycosylation; postfusion; spike; subtomogram averaging; vaccine; β-propiolactone-inactivated viruses.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing financial or non-financial interests.
Figures

References
-
- Castaño-Díez D., Kudryashev M., Arheit M., Stahlberg H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol. 2012;178:139–151. - PubMed
-
- Chen H., Xie Z., Long R., Fan S., Li H., He Z., Xu K., Liao Y., Wang L., Zhang Y. A valid protective immune response elicited in rhesus macaques by an inactivated vaccine is capable of defending against SARS-CoV-2 infection. bioRxiv. 2020
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

