Cancer-Associated Gain-of-Function Mutations Activate a SWI/SNF-Family Regulatory Hub
- PMID: 33058778
- PMCID: PMC7853424
- DOI: 10.1016/j.molcel.2020.09.024
Cancer-Associated Gain-of-Function Mutations Activate a SWI/SNF-Family Regulatory Hub
Abstract
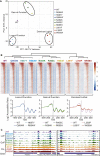
SWI/SNF-family remodelers (BAF/PBAF in mammals) are essential chromatin regulators, and mutations in human BAF/PBAF components are associated with ∼20% of cancers. Cancer-associated missense mutations in human BRG1 (encoding the catalytic ATPase) have been characterized previously as conferring loss-of-function. Here, we show that cancer-associated missense mutations in BRG1, when placed into the orthologous Sth1 ATPase of the yeast RSC remodeler, separate into two categories: loss-of-function enzymes, or instead, gain-of-function enzymes that greatly improve DNA translocation efficiency and nucleosome remodeling in vitro. Our work identifies a structural "hub," formed by the association of several Sth1 domains, that regulates ATPase activity and DNA translocation efficiency. Remarkably, all gain-of-function cancer-associated mutations and all loss-of-function mutations physically localize to distinct adjacent regions in the hub, which specifically regulate and implement DNA translocation, respectively. In vivo, only gain-of-function cancer-associated mutations conferred precocious chromatin accessibility. Taken together, we provide a structure-function mechanistic basis for cancer-associated hyperactivity.
Keywords: BAF; BRG1; DNA accessibility; RSC; STH1; SWI/SNF; cancer; chromatin remodeling; nucleosome.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

