The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis
- PMID: 33060131
- PMCID: PMC7710015
- DOI: 10.1242/dev.193862
The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis
Abstract
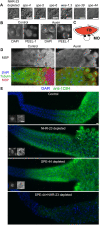
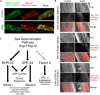
In sexually reproducing metazoans, spermatogenesis is the process by which uncommitted germ cells give rise to haploid sperm. Work in model systems has revealed mechanisms controlling commitment to the sperm fate, but how this fate is subsequently executed remains less clear. While studying the well-established role of the conserved nuclear hormone receptor transcription factor, NHR-23/NR1F1, in regulating C. elegans molting, we discovered that NHR-23/NR1F1 is also constitutively expressed in developing primary spermatocytes and is a critical regulator of spermatogenesis. In this novel role, NHR-23/NR1F1 functions downstream of the canonical sex-determination pathway. Degron-mediated depletion of NHR-23/NR1F1 within hermaphrodite or male germlines causes sterility due to an absence of functional sperm, as depleted animals produce arrested primary spermatocytes rather than haploid sperm. These spermatocytes arrest in prometaphase I and fail to either progress to anaphase or attempt spermatid-residual body partitioning. They make sperm-specific membranous organelles but fail to assemble their major sperm protein into fibrous bodies. NHR-23/NR1F1 appears to function independently of the known SPE-44 gene regulatory network, revealing the existence of an NHR-23/NR1F1-mediated module that regulates the spermatogenesis program.
Keywords: Auxin-inducible degron; C. elegans; Meiosis; NHR-23; Nuclear hormone receptor; Spermatogenesis.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
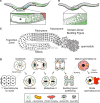
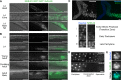

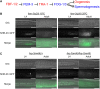

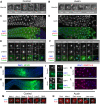
References
-
- Alvarez J. D., Hansen A., Ord T., Bebas P., Chappell P. E., Giebultowicz J. M., Williams C., Moss S. and Sehgal A. (2008). The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms 23, 26-36. 10.1177/0748730407311254 - DOI - PMC - PubMed
-
- André E., Conquet F., Steinmayr M., Stratton S. C., Porciatti V. and Becker-André M. (1998). Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867-3877. 10.1093/emboj/17.14.3867 - DOI - PMC - PubMed
-
- Ashley G., Duong T., Levenson M. T., Martinez M. A. Q., Hibshman J. D., Saeger H. N., Doonan R., Palmisano N. J., Martinez-Mendez R., Davidson B. et al. (2020). Expanding the Caenorhabditis elegans auxin-inducible degron system toolkit with internal expression and degradation controls and improved modular constructs for CRISPR/Cas9-mediated genome editing. bioRxiv. 10.1101/2020.05.12.090217 - DOI

