Health and disease markers correlate with gut microbiome composition across thousands of people
- PMID: 33060586
- PMCID: PMC7562722
- DOI: 10.1038/s41467-020-18871-1
Health and disease markers correlate with gut microbiome composition across thousands of people
Abstract
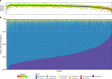
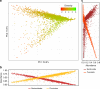
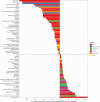
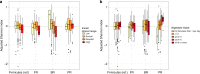
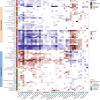
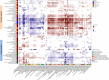
Variation in the human gut microbiome can reflect host lifestyle and behaviors and influence disease biomarker levels in the blood. Understanding the relationships between gut microbes and host phenotypes are critical for understanding wellness and disease. Here, we examine associations between the gut microbiota and ~150 host phenotypic features across ~3,400 individuals. We identify major axes of taxonomic variance in the gut and a putative diversity maximum along the Firmicutes-to-Bacteroidetes axis. Our analyses reveal both known and unknown associations between microbiome composition and host clinical markers and lifestyle factors, including host-microbe associations that are composition-specific. These results suggest potential opportunities for targeted interventions that alter the composition of the microbiome to improve host health. By uncovering the interrelationships between host diet and lifestyle factors, clinical blood markers, and the human gut microbiome at the population-scale, our results serve as a roadmap for future studies on host-microbe interactions and interventions.
Conflict of interest statement
O.M., C.L.D., S.A.K., B.S., J.C.L., and A.T.M. were all former employees and shareholders of Arivale. N.D.P. was a former shareholder of Arivale. Arivale is no longer a commercially operating company as of April 2019. The remaining author declares no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

