Genes affecting the extension of chronological lifespan in Schizosaccharomyces pombe (fission yeast)
- PMID: 33064911
- PMCID: PMC8246873
- DOI: 10.1111/mmi.14627
Genes affecting the extension of chronological lifespan in Schizosaccharomyces pombe (fission yeast)
Abstract
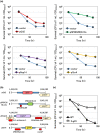
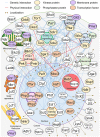
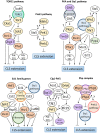
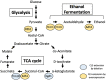
So far, more than 70 genes involved in the chronological lifespan (CLS) of Schizosaccharomyces pombe (fission yeast) have been reported. In this mini-review, we arrange and summarize these genes based on the reported genetic interactions between them and the physical interactions between their products. We describe the signal transduction pathways that affect CLS in S. pombe: target of rapamycin complex 1, cAMP-dependent protein kinase, Sty1, and Pmk1 pathways have important functions in the regulation of CLS extension. Furthermore, the Php transcription complex, Ecl1 family proteins, cyclin Clg1, and the cyclin-dependent kinase Pef1 are important for the regulation of CLS extension in S. pombe. Most of the known genes involved in CLS extension are related to these pathways and genes. In this review, we focus on the individual genes regulating CLS extension in S. pombe and discuss the interactions among them.
Keywords: Schizosaccharomyces pombe; chronological lifespan; fission yeast; longevity; signal transduction; stationary phase.
© 2020 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures
References
-
- Bicho, C.C. , de Lima Alves, F.D.L. , Chen, Z.A. , Rappsilber, J. and Sawin, K.E. (2010) A genetic engineering solution to the ‘arginine conversion problem’ in stable isotope labeling by amino acids in cell culture (SILAC). Molecular and Cellular Proteomics, 9, 1567–1577. 10.1074/mcp.M110.000208. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

