Seeding Biochemistry on Other Worlds: Enceladus as a Case Study
- PMID: 33064954
- PMCID: PMC7876360
- DOI: 10.1089/ast.2019.2197
Seeding Biochemistry on Other Worlds: Enceladus as a Case Study
Abstract
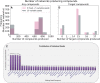
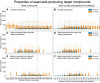
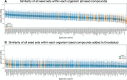
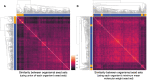
The Solar System is becoming increasingly accessible to exploration by robotic missions to search for life. However, astrobiologists currently lack well-defined frameworks to quantitatively assess the chemical space accessible to life in these alien environments. Such frameworks will be critical for developing concrete predictions needed for future mission planning, both to determine the potential viability of life on other worlds and to anticipate the molecular biosignatures that life could produce. Here, we describe how uniting existing methods provides a framework to study the accessibility of biochemical space across diverse planetary environments. Our approach combines observational data from planetary missions with genomic data catalogued from across Earth and analyzed using computational methods from network theory. To demonstrate this, we use 307 biochemical networks generated from genomic data collected across Earth and "seed" these networks with molecules confirmed to be present on Saturn's moon Enceladus. By expanding through known biochemical reaction space starting from these seed compounds, we are able to determine which products of Earth's biochemistry are, in principle, reachable from compounds available in the environment on Enceladus, and how this varies across different examples of life from Earth (organisms, ecosystems, planetary-scale biochemistry). While we find that none of the 307 prokaryotes analyzed meet the threshold for viability, the reaction space covered by this process can provide a map of possible targets for detection of Earth-like life on Enceladus, as well as targets for synthetic biology approaches to seed life on Enceladus. In cases where biochemistry is not viable because key compounds are missing, we identify the environmental precursors required to make it viable, thus providing a set of compounds to prioritize for detection in future planetary exploration missions aimed at assessing the ability of Enceladus to sustain Earth-like life or directed panspermia.
Keywords: Biochemical networks; Enceladus; Habitability; Life as a planetary process; Metabolic networks; Panspermia; Planetary protection.
Conflict of interest statement
No competing financial interests exist.
Figures



References
-
- Burdukiewicz M, Gagat P, Jablonski S, et al. (2018) PhyMet (2): a database and toolkit for phylogenetic and metabolic analyses of methanogens (vol 10, pg 378, 2018). Environ Microbiol Rep 10:605. - PubMed
-
- Cottret L, Milreu PV, Acuña V, et al. (2008) Enumerating precursor sets of target metabolites in a metabolic network. In International Workshop on Algorithms in Bioinformatics. Springer, Berlin; pp 233–244
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
