The epitranscriptome in stem cell biology and neural development
- PMID: 33065280
- PMCID: PMC7686257
- DOI: 10.1016/j.nbd.2020.105139
The epitranscriptome in stem cell biology and neural development
Abstract
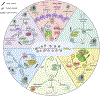
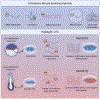
The blossoming field of epitranscriptomics has recently garnered attention across many fields by findings that chemical modifications on RNA have immense biological consequences. Methylation of nucleotides in RNA, including N6-methyladenosine (m6A), 2-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), 5-methylcytosine (m5C), and isomerization of uracil to pseudouridine (Ψ), have the potential to alter RNA processing events and contribute to developmental processes and different diseases. Though the abundance and roles of some RNA modifications remain contentious, the epitranscriptome is thought to be especially relevant in stem cell biology and neurobiology. In particular, m6A occurs at the highest levels in the brain and plays major roles in embryonic stem cell differentiation, brain development, and neurodevelopmental disorders. However, studies in these areas have reported conflicting results on epitranscriptomic regulation of stem cell pluripotency and mechanisms in neural development. In this review we provide an overview of the current understanding of several RNA modifications and disentangle the various findings on epitranscriptomic regulation of stem cell biology and neural development.
Keywords: Brain development; Brain disorders; Epitranscriptome; Stem cells; m(6)A.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

