Amplicon-Based Detection and Sequencing of SARS-CoV-2 in Nasopharyngeal Swabs from Patients With COVID-19 and Identification of Deletions in the Viral Genome That Encode Proteins Involved in Interferon Antagonism
- PMID: 33066701
- PMCID: PMC7602519
- DOI: 10.3390/v12101164
Amplicon-Based Detection and Sequencing of SARS-CoV-2 in Nasopharyngeal Swabs from Patients With COVID-19 and Identification of Deletions in the Viral Genome That Encode Proteins Involved in Interferon Antagonism
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). Sequencing the viral genome as the outbreak progresses is important, particularly in the identification of emerging isolates with different pathogenic potential and to identify whether nucleotide changes in the genome will impair clinical diagnostic tools such as real-time PCR assays. Although single nucleotide polymorphisms and point mutations occur during the replication of coronaviruses, one of the biggest drivers in genetic change is recombination. This can manifest itself in insertions and/or deletions in the viral genome. Therefore, sequencing strategies that underpin molecular epidemiology and inform virus biology in patients should take these factors into account. A long amplicon/read length-based RT-PCR sequencing approach focused on the Oxford Nanopore MinION/GridION platforms was developed to identify and sequence the SARS-CoV-2 genome in samples from patients with or suspected of COVID-19. The protocol, termed Rapid Sequencing Long Amplicons (RSLAs) used random primers to generate cDNA from RNA purified from a sample from a patient, followed by single or multiplex PCRs to generate longer amplicons of the viral genome. The base protocol was used to identify SARS-CoV-2 in a variety of clinical samples and proved sensitive in identifying viral RNA in samples from patients that had been declared negative using other nucleic acid-based assays (false negative). Sequencing the amplicons revealed that a number of patients had a proportion of viral genomes with deletions.
Keywords: MinION; SARS-CoV-2; amplicon; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

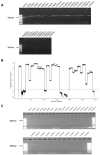

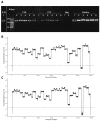
References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in SARS-CoV-2, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
-
- Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., Holden K.A., Read J.M., Dondelinger F., Carson G., et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. - DOI - PMC - PubMed
-
- Knight S.R., Ho A., Pius R., Buchan I., Carson G., Drake T.M., Dunning J., Fairfield C.J., Gamble C., Green C.A., et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

