Overexpression of an ethylene-forming ACC oxidase (ACO) gene precedes the Minute Hilum seed coat phenotype in Glycine max
- PMID: 33066734
- PMCID: PMC7566151
- DOI: 10.1186/s12864-020-07130-8
Overexpression of an ethylene-forming ACC oxidase (ACO) gene precedes the Minute Hilum seed coat phenotype in Glycine max
Abstract
Background: To elucidate features of seed development, we investigated the transcriptome of a soybean isoline from the germplasm collection that contained an introgressed allele known as minute hilum (mi) which confers a smaller hilum region where the seed attaches to the pod and also results in seed coat cracking surrounding the hilum region.
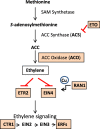
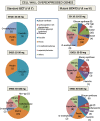
Results: RNAs were extracted from immature seed from an extended hilum region (i.e., the hilum and a small ring of tissue surrounding the hilum in which the cracks form) at three different developmental stages:10-25, 25-50 and 50-100 mg seed fresh weight in two independent replicates for each stage. The transcriptomes of these samples from both the Clark isoline containing the mi allele (PI 547628, UC413, ii R t mi G), and its recurrent Clark 63 parent isoline (PI 548532, UC7, ii R T Mi g), which was used for six generations of backcrossing, were compared for differential expression of 88,648 Glyma models of the soybean genome Wm82.a2. The RNA sequence data obtained from the 12 cDNA libraries were subjected to padj value < 0.05 and at least two-fold expression differences to select with confidence genes differentially expressed in the hilum-containing tissue of the seed coat between the two lines. Glyma.09G008400 annotated as encoding an ethylene forming enzyme, ACC oxidase (ACO), was found to be highly overexpressed in the mi hilum region at 165 RPKMs (reads per kilobase per million mapped reads) compared to the standard line at just 0.03 RPKMs. Evidence of changes in expression of genes downstream of the ethylene pathway included those involved in auxin and gibberellin hormone action and extensive differences in expression of cell wall protein genes. These changes are postulated to determine the restricted hilum size and cracking phenotypes.
Conclusions: We present transcriptome and phenotypic evidence that substantially higher expression of an ethylene-forming ACO gene likely shifts hormone balance and sets in motion downstream changes resulting in a smaller hilum phenotype and the cracks observed in the minute hilum (mi) isoline as compared to its recurrent parent.
Keywords: ACC oxidase; Defective seed coat; Ethylene forming enzyme; Expression profiling; Glycine max; Minute hilum; RNA-Seq; Seed development; Soybean.
Conflict of interest statement
The authors declare they have no competing interests.
Figures





Similar articles
-
RNA-Seq profiling of a defective seed coat mutation in Glycine max reveals differential expression of proline-rich and other cell wall protein transcripts.PLoS One. 2014 May 14;9(5):e96342. doi: 10.1371/journal.pone.0096342. eCollection 2014. PLoS One. 2014. PMID: 24828743 Free PMC article.
-
Loss-of-function mutations affecting a specific Glycine max R2R3 MYB transcription factor result in brown hilum and brown seed coats.BMC Plant Biol. 2011 Nov 9;11:155. doi: 10.1186/1471-2229-11-155. BMC Plant Biol. 2011. PMID: 22070454 Free PMC article.
-
Combined analysis of transcriptome and metabolite data reveals extensive differences between black and brown nearly-isogenic soybean (Glycine max) seed coats enabling the identification of pigment isogenes.BMC Genomics. 2011 Jul 29;12:381. doi: 10.1186/1471-2164-12-381. BMC Genomics. 2011. PMID: 21801362 Free PMC article.
-
The Ethylene Biosynthetic Enzymes, 1-Aminocyclopropane-1-Carboxylate (ACC) Synthase (ACS) and ACC Oxidase (ACO): The Less Explored Players in Abiotic Stress Tolerance.Biomolecules. 2024 Jan 11;14(1):90. doi: 10.3390/biom14010090. Biomolecules. 2024. PMID: 38254690 Free PMC article. Review.
-
Molecular mapping and genomics of soybean seed protein: a review and perspective for the future.Theor Appl Genet. 2017 Oct;130(10):1975-1991. doi: 10.1007/s00122-017-2955-8. Epub 2017 Aug 11. Theor Appl Genet. 2017. PMID: 28801731 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Functional Investigation of 1-Aminocyclopropane-1-carboxylic Acid Oxidase (ACO) Genes in Cotton.Plants (Basel). 2021 Aug 18;10(8):1699. doi: 10.3390/plants10081699. Plants (Basel). 2021. PMID: 34451744 Free PMC article.
-
Characterization of the Common Genetic Basis Underlying Seed Hilum Size, Yield, and Quality Traits in Soybean.Front Plant Sci. 2021 Feb 25;12:610214. doi: 10.3389/fpls.2021.610214. eCollection 2021. Front Plant Sci. 2021. PMID: 33719282 Free PMC article.
References
-
- Carlson JB, Lersten NR: Reproductive morphology. In Boerma HR, Specht JE, editors. Soybeans: improvement, production, and uses. Madison WI. 2004; American Society of Agronomy. pp. 59–95.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

