Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma
- PMID: 33067228
- PMCID: PMC7567591
- DOI: 10.1126/sciadv.abb5427
Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma
Abstract
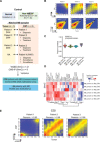
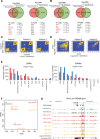
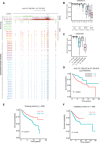
Medulloblastoma (MB), the most common form of pediatric brain malignancy, has a low frequency of oncogenic mutations but pronouncedly abnormal DNA methylation changes. Epigenetic analysis of circulating cell-free tumor DNA (ctDNA) by liquid biopsy offers an approach for real-time monitoring of tumor status without tumor dissection. In this study, we identified 6598 differentially methylated CpGs in both MB tumors and the ctDNA isolated from matched cerebrospinal fluid (CSF) compared with normal cerebellum, which could be used to detect MB tumor occurrence and determine its subtype. Furthermore, DNA methylation changes in serial CSF samples could be used to monitor the treatment response and tumor recurrence. Integrating our data with large public datasets, we identified reliable MB DNA methylation signatures in ctDNA that have potential diagnostic and prognostic values. Our study sets the stage for exploiting epigenetic markers in CSF to improve the clinical management of patients with MB.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Massimino M., Biassoni V., Gandola L., Garrè M. L., Gatta G., Giangaspero F., Poggi G., Rutkowski S., Childhood medulloblastoma. Crit. Rev. Oncol. Hematol. 105, 35–51 (2016). - PubMed
-
- Batora N. V., Sturm D., Jones D. T. W., Kool M., Pfister S. M., Northcott P. A., Transitioning from genotypes to epigenotypes: Why the time has come for medulloblastoma epigenomics. Neuroscience 264, 171–185 (2014). - PubMed
-
- Dubuc A. M., Remke M., Korshunov A., Northcott P. A., Zhan S. H., Mendez-Lago M., Kool M., Jones D. T. W., Unterberger A., Morrissy A. S., Shih D., Peacock J., Ramaswamy V., Rolider A., Wang X., Witt H., Hielscher T., Hawkins C., Vibhakar R., Croul S., Rutka J. T., Weiss W. A., Jones S. J. M., Eberhart C. G., Marra M. A., Pfister S. M., Taylor M. D., Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 125, 373–384 (2013). - PMC - PubMed
-
- Remke M., Ramaswamy V., Taylor M. D., Medulloblastoma molecular dissection: The way toward targeted therapy. Curr. Opin. Oncol. 25, 674–681 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

