SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans
- PMID: 33067384
- PMCID: PMC7569149
- DOI: 10.1128/mBio.01930-20
SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans
Abstract
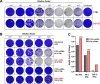
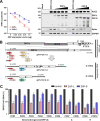
Recent evidence shows that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is sensitive to interferons (IFNs). However, the most effective types of IFNs and the underlying antiviral effectors remain to be defined. Here, we show that zinc finger antiviral protein (ZAP), which preferentially targets CpG dinucleotides in viral RNA sequences, restricts SARS-CoV-2. We further demonstrate that ZAP and its cofactors KHNYN and TRIM25 are expressed in human lung cells. Type I, II, and III IFNs all strongly inhibited SARS-CoV-2 and further induced ZAP expression. Comprehensive sequence analyses revealed that SARS-CoV-2 and its closest relatives from horseshoe bats showed the strongest CpG suppression among all known human and bat coronaviruses, respectively. Nevertheless, endogenous ZAP expression restricted SARS-CoV-2 replication in human lung cells, particularly upon treatment with IFN-α or IFN-γ. Both the long and the short isoforms of human ZAP reduced SARS-CoV-2 RNA expression levels, but the former did so with greater efficiency. Finally, we show that the ability to restrict SARS-CoV-2 is conserved in ZAP orthologues of the reservoir bat and potential intermediate pangolin hosts of human coronaviruses. Altogether, our results show that ZAP is an important effector of the innate response against SARS-CoV-2, although this pandemic pathogen emerged from zoonosis of a coronavirus that was preadapted to the low-CpG environment in humans.IMPORTANCE Although interferons inhibit SARS-CoV-2 and have been evaluated for treatment of coronavirus disease 2019 (COVID-19), the most effective types and antiviral effectors remain to be defined. Here, we show that IFN-γ is particularly potent in restricting SARS-CoV-2 and in inducing expression of the antiviral factor ZAP in human lung cells. Knockdown experiments revealed that endogenous ZAP significantly restricts SARS-CoV-2. We further show that CpG dinucleotides which are specifically targeted by ZAP are strongly suppressed in the SARS-CoV-2 genome and that the two closest horseshoe bat relatives of SARS-CoV-2 show the lowest genomic CpG content of all coronavirus sequences available from this reservoir host. Nonetheless, both the short and long isoforms of human ZAP reduced SARS-CoV-2 RNA levels, and this activity was conserved in horseshoe bat and pangolin ZAP orthologues. Our findings indicating that type II interferon is particularly efficient against SARS-CoV-2 and that ZAP restricts this pandemic viral pathogen might promote the development of effective immune therapies against COVID-19.
Keywords: COVID-19; CpG suppression; SARS-CoV-2; ZAP; evolution; interferon.
Copyright © 2020 Nchioua et al.
Figures





References
-
- Zhou P, Yang X-L, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang R-D, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
