Non-invasive decision support for NSCLC treatment using PET/CT radiomics
- PMID: 33067442
- PMCID: PMC7567795
- DOI: 10.1038/s41467-020-19116-x
Non-invasive decision support for NSCLC treatment using PET/CT radiomics
Abstract
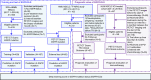
Two major treatment strategies employed in non-small cell lung cancer, NSCLC, are tyrosine kinase inhibitors, TKIs, and immune checkpoint inhibitors, ICIs. The choice of strategy is based on heterogeneous biomarkers that can dynamically change during therapy. Thus, there is a compelling need to identify comprehensive biomarkers that can be used longitudinally to help guide therapy choice. Herein, we report a 18F-FDG-PET/CT-based deep learning model, which demonstrates high accuracy in EGFR mutation status prediction across patient cohorts from different institutions. A deep learning score (EGFR-DLS) was significantly and positively associated with longer progression free survival (PFS) in patients treated with EGFR-TKIs, while EGFR-DLS is significantly and negatively associated with higher durable clinical benefit, reduced hyperprogression, and longer PFS among patients treated with ICIs. Thus, the EGFR-DLS provides a non-invasive method for precise quantification of EGFR mutation status in NSCLC patients, which is promising to identify NSCLC patients sensitive to EGFR-TKI or ICI-treatments.
Conflict of interest statement
R.J.G. declared a potential conflict with HealthMyne, Inc (Investor (major), Board of Advisors (uncompensated)). J.E.G. reports receiving commercial research grants from AstraZeneca, Merck, Array, Epic Sciences, Genentech, Bristol-Myers Squibb, BI, Trovagene, and Novartis, and is a consultant/advisory board member for AstraZeneca, Janssen, Genentech, Eli Lilly, Celgene, and Takeda, and other remuneration from Genentech, AstraZeneca, Merck, and Lilly/Genenech. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

