Identification of plastic-associated species in the Mediterranean Sea using DNA metabarcoding with Nanopore MinION
- PMID: 33067509
- PMCID: PMC7568539
- DOI: 10.1038/s41598-020-74180-z
Identification of plastic-associated species in the Mediterranean Sea using DNA metabarcoding with Nanopore MinION
Abstract
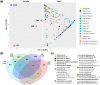
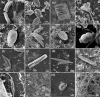
Plastic debris in the ocean form a new ecosystem, termed 'plastisphere', which hosts a variety of marine organisms. Recent studies implemented DNA metabarcoding to characterize the taxonomic composition of the plastisphere in different areas of the world. In this study, we used a modified metabarcoding approach which was based on longer barcode sequences for the characterization of the plastisphere biota. We compared the microbiome of polyethylene food bags after 1 month at sea to the free-living biome in two proximal but environmentally different locations on the Mediterranean coast of Israel. We targeted the full 1.5 kb-long 16S rRNA gene for bacteria and 0.4-0.8 kb-long regions within the 18S rRNA, ITS, tufA and COI loci for eukaryotes. The taxonomic barcodes were sequenced using Oxford Nanopore Technology with multiplexing on a single MinION flow cell. We identified between 1249 and 2141 species in each of the plastic samples, of which 61 species (34 bacteria and 27 eukaryotes) were categorized as plastic-specific, including species that belong to known hydrocarbon-degrading genera. In addition to a large prokaryotes repertoire, our results, supported by scanning electron microscopy, depict a surprisingly high biodiversity of eukaryotes within the plastisphere with a dominant presence of diatoms as well as other protists, algae and fungi.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Boucher J, Billard G. The challenges of measuring plastic pollution. Field Actions Sci. Rep. J. Field Actions. 2019;19:68–75.
-
- Worm B, Lotze HK, Jubinville I, Wilcox C, Jambeck J. Plastic as a persistent marine pollutant. Annu. Rev. Environ. Resour. 2017;42:1–26. doi: 10.1146/annurev-environ-102016-060700. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

