Development of a prognostic model for mortality in COVID-19 infection using machine learning
- PMID: 33067522
- PMCID: PMC7567420
- DOI: 10.1038/s41379-020-00700-x
Development of a prognostic model for mortality in COVID-19 infection using machine learning
Abstract
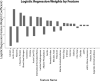
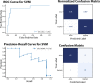
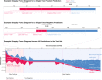
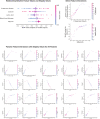
Coronavirus disease 2019 (COVID-19) is a novel disease resulting from infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has quickly risen since the beginning of 2020 to become a global pandemic. As a result of the rapid growth of COVID-19, hospitals are tasked with managing an increasing volume of these cases with neither a known effective therapy, an existing vaccine, nor well-established guidelines for clinical management. The need for actionable knowledge amidst the COVID-19 pandemic is dire and yet, given the urgency of this illness and the speed with which the healthcare workforce must devise useful policies for its management, there is insufficient time to await the conclusions of detailed, controlled, prospective clinical research. Thus, we present a retrospective study evaluating laboratory data and mortality from patients with positive RT-PCR assay results for SARS-CoV-2. The objective of this study is to identify prognostic serum biomarkers in patients at greatest risk of mortality. To this end, we develop a machine learning model using five serum chemistry laboratory parameters (c-reactive protein, blood urea nitrogen, serum calcium, serum albumin, and lactic acid) from 398 patients (43 expired and 355 non-expired) for the prediction of death up to 48 h prior to patient expiration. The resulting support vector machine model achieved 91% sensitivity and 91% specificity (AUC 0.93) for predicting patient expiration status on held-out testing data. Finally, we examine the impact of each feature and feature combination in light of different model predictions, highlighting important patterns of laboratory values that impact outcomes in SARS-CoV-2 infection.
Figures
References
-
- Johns Hopkins University & Medicine. Johns Hopkins Coronavirus Resource Center. 2020. https://coronavirus.jhu.edu/.
-
- Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7146693/. - PMC - PubMed
-
- Zhou B, She J, Wang Y, Ma X. Utility of ferritin, procalcitonin, and c-reactive protein in severe patients with 2019 novel coronavirus disease. 2020. https://www.researchsquare.com/article/rs-18079/v1.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

