Characterization of Metabolic Patterns in Mouse Oocytes during Meiotic Maturation
- PMID: 33068521
- PMCID: PMC8034554
- DOI: 10.1016/j.molcel.2020.09.022
Characterization of Metabolic Patterns in Mouse Oocytes during Meiotic Maturation
Abstract
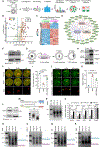
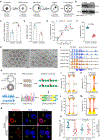
Well-balanced and timed metabolism is essential for making a high-quality egg. However, the metabolic framework that supports oocyte development remains poorly understood. Here, we obtained the temporal metabolome profiles of mouse oocytes during in vivo maturation by isolating large number of cells at key stages. In parallel, quantitative proteomic analyses were conducted to bolster the metabolomic data, synergistically depicting the global metabolic patterns in oocytes. In particular, we discovered the metabolic features during meiotic maturation, such as the fall in polyunsaturated fatty acids (PUFAs) level and the active serine-glycine-one-carbon (SGOC) pathway. Using functional approaches, we further identified the key targets mediating the action of PUFA arachidonic acid (ARA) on meiotic maturation and demonstrated the control of epigenetic marks in maturing oocytes by SGOC network. Our data serve as a broad resource on the dynamics occurring in metabolome and proteome during oocyte maturation.
Keywords: embryo; epigenetics; metabolism; oocyte; proteomics.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Bodden C, Hannan AJ, and Reichelt AC (2020). Diet-Induced Modification of the Sperm Epigenome Programs Metabolism and Behavior. Trends Endocrinol Metab 31, 131–149. - PubMed
-
- Broughton DE, and Moley KH (2017). Obesity and Female Infertility: Potential Mediators of Obesity’s Impact. Fertil Steril 107, 840–847. - PubMed
-
- Brunet S, Pahlavan G, Taylor S, and Maro B (2003). Functionality of the Spindle Checkpoint During the First Meiotic Division of Mammalian Oocytes. Reproduction 126, 443–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

