Specificity and positive predictive value of SARS-CoV-2 nucleic acid amplification testing in a low-prevalence setting
- PMID: 33068757
- PMCID: PMC7554481
- DOI: 10.1016/j.cmi.2020.10.003
Specificity and positive predictive value of SARS-CoV-2 nucleic acid amplification testing in a low-prevalence setting
Abstract
Objectives: When the prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is low, many positive test results are false positives. Confirmatory testing reduces overdiagnosis and nosocomial infection and enables real-world estimates of test specificity and positive predictive value. This study estimates these parameters to evaluate the impact of confirmatory testing and to improve clinical diagnosis, epidemiological estimation and interpretation of vaccine trials.
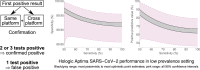
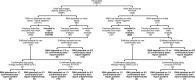
Methods: Over 1 month we took all respiratory samples from our laboratory with a patient's first detection of SARS-CoV-2 RNA (Hologic Aptima SARS-CoV-2 assay or in-house RT-PCR platform), and repeated testing using two platforms. Samples were categorized by source, and by whether clinical details suggested COVID-19 or corroborative testing from another laboratory. We estimated specificity and positive predictive value using approaches based on maximum likelihood.
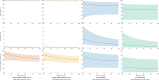
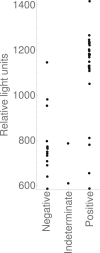
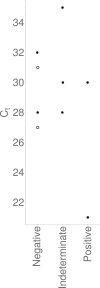
Results: Of 19 597 samples, SARS-CoV-2 RNA was detected in 107; 52 corresponded to first-time detection (0.27% of tests on samples without previous detection). Further testing detected SARS-CoV-2 RNA once or more ('confirmed') in 29 samples (56%), and failed to detect SARS-CoV-2 RNA ('not confirmed') in 23 (44%). Depending upon assumed parameters, point estimates for specificity and positive predictive value were 99.91-99.98% and 61.8-89.8% respectively using the Hologic Aptima SARS-CoV-2 assay, and 97.4-99.1% and 20.1-73.8% respectively using an in-house assay.
Conclusions: Nucleic acid amplification testing for SARS-CoV-2 is highly specific. Nevertheless, when prevalence is low a significant proportion of initially positive results fail to confirm, and confirmatory testing substantially reduces the detection of false positives. Omitting additional testing in samples with higher prior detection probabilities focuses testing where it is clinically impactful and minimizes delay.
Keywords: COVID-19 diagnostic testing; COVID-19 pandemic; Nucleic acid amplification techniques; SARS-CoV-2; Sensitivity and specificity.
Copyright © 2020 European Society of Clinical Microbiology and Infectious Diseases. All rights reserved.
Figures
References
-
- Nogueirai P.J., De Araújo Nobre M., Nicola P.J., Furtado C., Vaz Carneiro A. Excess mortality estimation during the COVID-19 pandemic: preliminary data from Portugal. Acta Med Port. 2020;33:376–383. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

