Interaction kinetics of peptide lipids-mediated gene delivery
- PMID: 33069258
- PMCID: PMC7568367
- DOI: 10.1186/s12951-020-00707-1
Interaction kinetics of peptide lipids-mediated gene delivery
Abstract
Background: During the course of gene transfection, the interaction kinetics between liposomes and DNA is speculated to play very important role for blood stability, cellular uptake, DNA release and finally transfection efficiency.
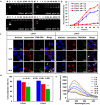
Results: As cationic peptide liposomes exhibited great gene transfer activities both in vitro and in vivo, two peptide lipids, containing a tri-ornithine head (LOrn3) and a mono-ornithine head (LOrn1), were chosen to further clarify the process of liposome-mediated gene delivery in this study. The results show that the electrostatically-driven binding between DNA and liposomes reached nearly 100% at equilibrium, and high affinity of LOrn3 to DNA led to fast binding rate between them. The binding process between LOrn3 and DNA conformed to the kinetics equation: y = 1.663631 × exp (- 0.003427x) + 6.278163. Compared to liposome LOrn1, the liposome LOrn3/DNA lipoplex exhibited a faster and more uniform uptake in HeLa cells, as LOrn3 with a tri-ornithine peptide headgroup had a stronger interaction with the negatively charged cell membrane than LOrn1. The efficient endosomal escape of DNA from LOrn3 lipoplex was facilitated by the acidity in late endosomes, resulting in broken carbamate bonds, as well as the "proton sponge effect" of the lipid.
Conclusions: The interaction kinetics is a key factor for DNA transfection efficiency. This work provided insights into peptide lipid-mediated DNA delivery that could guide the development of the next generation of delivery systems for gene therapeutics.
Keywords: Gene delivery; Gene release; Interaction kinetics; Peptide lipids; Uptake.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Niu J, Chu Y, Huang YF, Chong YS, Jiang ZH, Mao ZW, Peng LH, Gao JQ. Transdermal gene delivery by functional peptide-conjugated cationic gold nanoparticle reverses the progression and metastasis of cutaneous melanoma. ACS Appl Mater Interfaces. 2017;9:9388–9401. doi: 10.1021/acsami.6b16378. - DOI - PubMed
-
- Ashley CE, Carnes EC, Epler KE, Padilla DP, Phillips GK, Castillo RE, Wilkinson DC, Wilkinson BS, Burgard CA, Kalinich RM, Townson JL, Chackerian B, Willman CL, Peabody DS, Wharton W, Brinker CJ. Delivery of small interfering RNA by peptide-targeted mesoporous silica nanoparticle-supported lipid bilayers. ACS Nano. 2012;3:2174–2188. doi: 10.1021/nn204102q. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

