Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies
- PMID: 33069360
- PMCID: PMC7547570
- DOI: 10.1016/j.bbrc.2020.10.012
Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies
Abstract
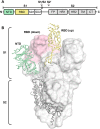
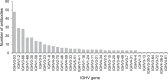
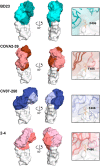
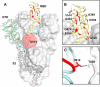
Immediately from the outset of the COVID-19 pandemic, researchers from diverse biomedical and biological disciplines have united to study the novel pandemic virus, SARS-CoV-2. The antibody response to SARS-CoV-2 has been a major focus of COVID-19 research due to its clinical relevance and importance in vaccine and therapeutic development. Isolation and characterization of antibodies to SARS-CoV-2 have been accumulating at an unprecedented pace. Most of the SARS-CoV-2 neutralizing antibodies to date target the spike (S) protein receptor binding domain (RBD), which engages the host receptor ACE2 for viral entry. Here we review the binding sites and molecular features of monoclonal antibodies that target the SARS-CoV-2 RBD, including a few that also cross-neutralize SARS-CoV.
Keywords: Antibody avidity; Cross-neutralization; Epitopes; Germline-encoded motifs; Neutralizing antibodies; RBD natural mutations; Receptor binding domain (RBD); SARS-CoV; SARS-CoV-2.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- WHO Novel coronavirus – China. Jan 12, 2020. 2020. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc8413. - DOI - PMC - PubMed
-
- Han X., Wang Y., Li S., Hu C., Li T., Gu C., Wang K., Shen M., Wang J., Hu J., Wu R., Mu S., Gong F., Chen Q., Gao F., Huang J., Long Y., Luo F., Song S., Long S., Hao Y., Li L., Wu Y., Xu W., Cai X., Gao Q., Zhang G., He C., Deng K., Du L., Nai Y., Wang W., Xie Y., Qu D., Huang A., Tang N., Jin A. bioRxiv; 2020. A Rapid and Efficient Screening System for Neutralizing Antibodies and its Application for the Discovery of Potent Neutralizing Antibodies to SARS-CoV-2 S-RBD. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

