The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions
- PMID: 33070389
- PMCID: PMC7737760
- DOI: 10.1002/pro.3978
The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions
Abstract
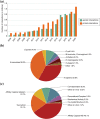
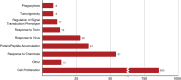
The BioGRID (Biological General Repository for Interaction Datasets, thebiogrid.org) is an open-access database resource that houses manually curated protein and genetic interactions from multiple species including yeast, worm, fly, mouse, and human. The ~1.93 million curated interactions in BioGRID can be used to build complex networks to facilitate biomedical discoveries, particularly as related to human health and disease. All BioGRID content is curated from primary experimental evidence in the biomedical literature, and includes both focused low-throughput studies and large high-throughput datasets. BioGRID also captures protein post-translational modifications and protein or gene interactions with bioactive small molecules including many known drugs. A built-in network visualization tool combines all annotations and allows users to generate network graphs of protein, genetic and chemical interactions. In addition to general curation across species, BioGRID undertakes themed curation projects in specific aspects of cellular regulation, for example the ubiquitin-proteasome system, as well as specific disease areas, such as for the SARS-CoV-2 virus that causes COVID-19 severe acute respiratory syndrome. A recent extension of BioGRID, named the Open Repository of CRISPR Screens (ORCS, orcs.thebiogrid.org), captures single mutant phenotypes and genetic interactions from published high throughput genome-wide CRISPR/Cas9-based genetic screens. BioGRID-ORCS contains datasets for over 1,042 CRISPR screens carried out to date in human, mouse and fly cell lines. The biomedical research community can freely access all BioGRID data through the web interface, standardized file downloads, or via model organism databases and partner meta-databases.
Keywords: COVID-19; CRISPR screen; biological network; chemical interaction; drug target; genetic interaction; phenotype; post-translational modification; protein interaction; ubiquitin-proteasome system.
© 2020 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

