Genome-enabled insights into the biology of thrips as crop pests
- PMID: 33070780
- PMCID: PMC7570057
- DOI: 10.1186/s12915-020-00862-9
Genome-enabled insights into the biology of thrips as crop pests
Erratum in
-
Correction to: Genome-enabled insights into the biology of thrips as crop pests.BMC Biol. 2020 Nov 16;18(1):169. doi: 10.1186/s12915-020-00915-z. BMC Biol. 2020. PMID: 33198778 Free PMC article.
Abstract
Background: The western flower thrips, Frankliniella occidentalis (Pergande), is a globally invasive pest and plant virus vector on a wide array of food, fiber, and ornamental crops. The underlying genetic mechanisms of the processes governing thrips pest and vector biology, feeding behaviors, ecology, and insecticide resistance are largely unknown. To address this gap, we present the F. occidentalis draft genome assembly and official gene set.
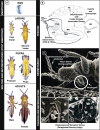
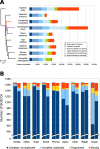
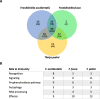
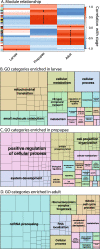
Results: We report on the first genome sequence for any member of the insect order Thysanoptera. Benchmarking Universal Single-Copy Ortholog (BUSCO) assessments of the genome assembly (size = 415.8 Mb, scaffold N50 = 948.9 kb) revealed a relatively complete and well-annotated assembly in comparison to other insect genomes. The genome is unusually GC-rich (50%) compared to other insect genomes to date. The official gene set (OGS v1.0) contains 16,859 genes, of which ~ 10% were manually verified and corrected by our consortium. We focused on manual annotation, phylogenetic, and expression evidence analyses for gene sets centered on primary themes in the life histories and activities of plant-colonizing insects. Highlights include the following: (1) divergent clades and large expansions in genes associated with environmental sensing (chemosensory receptors) and detoxification (CYP4, CYP6, and CCE enzymes) of substances encountered in agricultural environments; (2) a comprehensive set of salivary gland genes supported by enriched expression; (3) apparent absence of members of the IMD innate immune defense pathway; and (4) developmental- and sex-specific expression analyses of genes associated with progression from larvae to adulthood through neometaboly, a distinct form of maturation differing from either incomplete or complete metamorphosis in the Insecta.
Conclusions: Analysis of the F. occidentalis genome offers insights into the polyphagous behavior of this insect pest that finds, colonizes, and survives on a widely diverse array of plants. The genomic resources presented here enable a more complete analysis of insect evolution and biology, providing a missing taxon for contemporary insect genomics-based analyses. Our study also offers a genomic benchmark for molecular and evolutionary investigations of other Thysanoptera species.
Keywords: Chemosensory receptors; Detoxification; Hemipteroid assemblage; Innate immunity; Insect genomics; Opsins; Salivary glands; Thysanoptera; Tospovirus; Western flower thrips.
Conflict of interest statement
The authors declare they have no competing interests.
Figures


References
-
- Mound LA, Heming BS, Palmer JM. Phylogenetic relationships between the families of recent Thysanoptera (Insecta) Zool J Linn Soc. 1980;69:111–141.
-
- Ullman DE, Sherwood JL, German TL. Thrips as vectors of plant pathogens. In: Lewis TS, editor. Thrips as Crop Pests. Cambridge: CAB International; 1997. p. 539–65.
-
- Ullman DE, Wescot DM, Hunter WB, Mau RFL. Internal anatomy and morphology of Frankliniella occidentalis (Pergande) (Thysanoptera:Thripidae) with special reference to interactions between thrips and Tomato spotted wilt virus. Int J Insect Morphol Embryol. 1989;18:289–310.
-
- Ullman DE, Cho JJ, Mau RFL, Hunter WB, Westcot DM, Custer DM. Thrips-Tomato spotted wilt virus interactions: morphological, behavioral and cellular components influencing thrips transmission. In: Harris KF, editor. Advances in Disease Vector Research. New York: Springer-Verlag; 1992. pp. 195–240.
-
- Hunter WB, Ullman DE. Analysis of mouthpart movements during feeding of Frankliniella occidentalis (Pergande) and Frankliniella schultzei Trybom (Thysanoptera: Thripidae) Int. J. Insect Morphol. Embryol. 1989;18(2–3):161–172.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

