New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts
- PMID: 33071966
- PMCID: PMC7541902
- DOI: 10.3389/fendo.2020.538196
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts
Erratum in
-
Corrigendum: New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts.Front Endocrinol (Lausanne). 2020 Oct 28;11:610274. doi: 10.3389/fendo.2020.610274. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33193114 Free PMC article.
Abstract
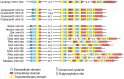
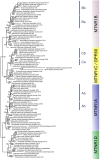
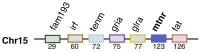
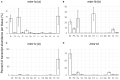
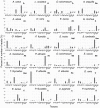
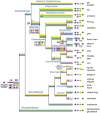
In order to improve our understanding of melatonin signaling, we have reviewed and revised the evolutionary history of melatonin receptor genes (mtnr) in vertebrates. All gnathostome mtnr genes have a conserved gene organization with two exons, except for mtnr1b paralogs of some teleosts that show intron gains. Phylogeny and synteny analyses demonstrate the presence of four mtnr subtypes, MTNR1A, MTNR1B, MTNR1C, MTNR1D that arose from duplication of an ancestral mtnr during the vertebrate tetraploidizations (1R and 2R). In tetrapods, mtnr1d was lost, independently, in mammals, in archosaurs and in caecilian amphibians. All four mtnr subtypes were found in two non-teleost actinopterygian species, the spotted gar and the reedfish. As a result of teleost tetraploidization (3R), up to seven functional mtnr genes could be identified in teleosts. Conservation of the mtnr 3R-duplicated paralogs differs among the teleost lineages. Synteny analysis showed that the mtnr1d was conserved as a singleton in all teleosts resulting from an early loss after tetraploidization of one of the teleost 3R and salmonid 4R paralogs. Several teleosts including the eels and the piranha have conserved both 3R-paralogs of mtnr1a, mtnr1b, and mtnr1c. Loss of one of the 3R-paralogs depends on the lineage: mtnr1ca was lost in euteleosts whereas mtnr1cb was lost in osteoglossomorphs and several ostariophysians including the zebrafish. We investigated the tissue distribution of mtnr expression in a large range of tissues in medaka. The medaka has conserved the four vertebrate paralogs, and these are expressed in brain and retina, and, differentially, in peripheral tissues. Photoperiod affects mtnr expression levels in a gene-specific and tissue-specific manner. This study provides new insights into the repertoire diversification and functional evolution of the mtnr gene family in vertebrates.
Keywords: functional evolution; gene duplication; medaka; melatonin receptors; phylogeny; synteny; teleosts; vertebrates.
Copyright © 2020 Maugars, Nourizadeh-Lillabadi and Weltzien.
Figures



Similar articles
-
Vertebrates originally possess four functional subtypes of G protein-coupled melatonin receptor.Sci Rep. 2019 Jul 1;9(1):9465. doi: 10.1038/s41598-019-45925-2. Sci Rep. 2019. PMID: 31263128 Free PMC article.
-
Basal teleosts provide new insights into the evolutionary history of teleost-duplicated aromatase.Gen Comp Endocrinol. 2020 May 15;291:113395. doi: 10.1016/j.ygcen.2020.113395. Epub 2020 Jan 23. Gen Comp Endocrinol. 2020. PMID: 31981691
-
New Insights Into the Evolution of Corticotropin-Releasing Hormone Family With a Special Focus on Teleosts.Front Endocrinol (Lausanne). 2022 Jul 22;13:937218. doi: 10.3389/fendo.2022.937218. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35937826 Free PMC article.
-
New insights into the evolution of vertebrate CRH (corticotropin-releasing hormone) and invertebrate DH44 (diuretic hormone 44) receptors in metazoans.Gen Comp Endocrinol. 2014 Dec 1;209:162-70. doi: 10.1016/j.ygcen.2014.09.004. Epub 2014 Sep 16. Gen Comp Endocrinol. 2014. PMID: 25230393 Review.
-
Evolutionary context can clarify gene names: Teleosts as a case study.Bioessays. 2021 Jun;43(6):e2000258. doi: 10.1002/bies.202000258. Epub 2021 Apr 7. Bioessays. 2021. PMID: 33829511 Review.
Cited by
-
The melatonin receptor 1B gene links circadian rhythms and type 2 diabetes mellitus: an evolutionary story.Ann Med. 2023 Dec;55(1):1262-1286. doi: 10.1080/07853890.2023.2191218. Ann Med. 2023. PMID: 36974476 Free PMC article. Review.
-
Pharmacological modulation of MRAP2 protein on murine melatonin receptor signaling.Front Endocrinol (Lausanne). 2025 May 29;16:1593345. doi: 10.3389/fendo.2025.1593345. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40510472 Free PMC article.
-
Day length regulates gonadotrope proliferation and reproduction via an intra-pituitary pathway in the model vertebrate Oryzias latipes.Commun Biol. 2024 Mar 30;7(1):388. doi: 10.1038/s42003-024-06059-y. Commun Biol. 2024. PMID: 38553567 Free PMC article.
-
Seasonal expression of reproductive axis-related neuroendocrine genes and their relation with ovarian maturation in captive yellowtail kingfish (Seriola lalandi).Biol Res. 2025 Aug 8;58(1):55. doi: 10.1186/s40659-025-00622-5. Biol Res. 2025. PMID: 40775662 Free PMC article.
-
Nocturnal melatonin increases glucose uptake via insulin-independent action in the goldfish brain.Front Endocrinol (Lausanne). 2023 May 23;14:1173113. doi: 10.3389/fendo.2023.1173113. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37288290 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

