Single Cell Genomics Reveals Viruses Consumed by Marine Protists
- PMID: 33072003
- PMCID: PMC7541821
- DOI: 10.3389/fmicb.2020.524828
Single Cell Genomics Reveals Viruses Consumed by Marine Protists
Abstract
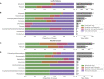
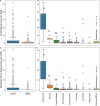
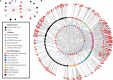
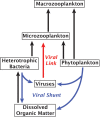
The predominant model of the role of viruses in the marine trophic web is that of the "viral shunt," where viral infection funnels a substantial fraction of the microbial primary and secondary production back to the pool of dissolved organic matter. Here, we analyzed the composition of non-eukaryotic DNA associated with individual cells of small, planktonic protists in the Gulf of Maine (GoM) and the Mediterranean Sea. We found viral DNA associated with a substantial fraction cells from the GoM (51%) and the Mediterranean Sea (35%). While Mediterranean SAGs contained a larger proportion of cells containing bacterial sequences (49%), a smaller fraction of cells contained bacterial sequences in the GoM (19%). In GoM cells, nearly identical bacteriophage and ssDNA virus sequences where found across diverse lineages of protists, suggesting many of these viruses are non-infective. The fraction of cells containing viral DNA varied among protistan lineages and reached 100% in Picozoa and Choanozoa. These two groups also contained significantly higher numbers of viral sequences than other identified taxa. We consider mechanisms that may explain the presence of viral DNA in protistan cells and conclude that protistan predation on free viral particles contributed to the observed patterns. These findings confirm prior experiments with protistan isolates and indicate that the viral shunt is complemented by a viral link in the marine microbial food web. This link may constitute a sink of viral particles in the ocean and has implications for the flow of carbon through the microbial food web.
Keywords: marine eukaryote; marine food web; microbial ecology; nanoeukaryote; phage; protist; virus.
Copyright © 2020 Brown, Labonté, Brown, Record, Poulton, Sieracki, Logares and Stepanauskas.
Figures
References
-
- Azam F., Fenchel T., Field J. G., Gray J., Meyer-Reil L., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10 257–263. 10.3354/meps010257 - DOI
-
- Bennett R. M., Honda D., Beakes G. W., Thines M. (2017). “Labyrinthulomycota,” in Handbook of the Protists, eds Archibald J. M., Simpson A. G. B., Slamovits C. H., Margulis L., Melkonian M., Chapman D. J., et al. (Cham: Springer International Publishing; ), 1–36. 10.1007/978-3-319-32669-6_25-1 - DOI
-
- Bettarel Y., Sime-Ngando T., Bouvy M., Arfi R., Amblard C. (2005). Low consumption of virus-sized particles by heterotrophic nanoflagellates in two lakes of the French Massif Central. Aquat. Microb. Ecol. 39 205–209. 10.3354/ame039205 - DOI
LinkOut - more resources
Full Text Sources

