Lineage-Specific Proteomic Signatures in the Mycobacterium tuberculosis Complex Reveal Differential Abundance of Proteins Involved in Virulence, DNA Repair, CRISPR-Cas, Bioenergetics and Lipid Metabolism
- PMID: 33072011
- PMCID: PMC7536270
- DOI: 10.3389/fmicb.2020.550760
Lineage-Specific Proteomic Signatures in the Mycobacterium tuberculosis Complex Reveal Differential Abundance of Proteins Involved in Virulence, DNA Repair, CRISPR-Cas, Bioenergetics and Lipid Metabolism
Abstract
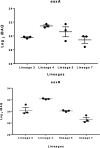
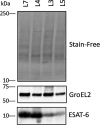
Despite the discovery of the tubercle bacillus more than 130 years ago, its physiology and the mechanisms of virulence are still not fully understood. A comprehensive analysis of the proteomes of members of the human-adapted Mycobacterium tuberculosis complex (MTBC) lineages 3, 4, 5, and 7 was conducted to better understand the evolution of virulence and other physiological characteristics. Unique and shared proteomic signatures in these modern, pre-modern and ancient MTBC lineages, as deduced from quantitative bioinformatics analyses of high-resolution mass spectrometry data, were delineated. The main proteomic findings were verified by using immunoblotting. In addition, analysis of multiple genome alignment of members of the same lineages was performed. Label-free peptide quantification of whole cells from MTBC lineages 3, 4, 5, and 7 yielded a total of 38,346 unique peptides derived from 3092 proteins, representing 77% coverage of the predicted proteome. MTBC lineage-specific differential expression was observed for 539 proteins. Lineage 7 exhibited a markedly reduced abundance of proteins involved in DNA repair, type VII ESX-3 and ESX-1 secretion systems, lipid metabolism and inorganic phosphate uptake, and an increased abundance of proteins involved in alternative pathways of the TCA cycle and the CRISPR-Cas system as compared to the other lineages. Lineages 3 and 4 exhibited a higher abundance of proteins involved in virulence, DNA repair, drug resistance and other metabolic pathways. The high throughput analysis of the MTBC proteome by super-resolution mass spectrometry provided an insight into the differential expression of proteins between MTBC lineages 3, 4, 5, and 7 that may explain the slow growth and reduced virulence, metabolic flexibility, and the ability to survive under adverse growth conditions of lineage 7.
Keywords: DNA repair; ESX-3 secretion system; Ethiopia; Mycobacterium tuberculosis; lineage 7; proteomics; virulence.
Copyright © 2020 Yimer, Kalayou, Homberset, Birhanu, Riaz, Zegeye, Lutter, Abebe, Holm-Hansen, Aseffa and Tønjum.
Figures



Similar articles
-
Comparative Proteomic Analysis of Mycobacterium tuberculosis Lineage 7 and Lineage 4 Strains Reveals Differentially Abundant Proteins Linked to Slow Growth and Virulence.Front Microbiol. 2017 May 9;8:795. doi: 10.3389/fmicb.2017.00795. eCollection 2017. Front Microbiol. 2017. PMID: 28536560 Free PMC article.
-
Identification of Quantitative Proteomic Differences between Mycobacterium tuberculosis Lineages with Altered Virulence.Front Microbiol. 2016 May 31;7:813. doi: 10.3389/fmicb.2016.00813. eCollection 2016. Front Microbiol. 2016. PMID: 27303394 Free PMC article.
-
The Bioinformatics Analysis of Comparative Genomics of Mycobacterium tuberculosis Complex (MTBC) Provides Insight into Dissimilarities between Intraspecific Groups Differing in Host Association, Virulence, and Epitope Diversity.Front Cell Infect Microbiol. 2017 Mar 21;7:88. doi: 10.3389/fcimb.2017.00088. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28377903 Free PMC article.
-
Lineage-specific differences in lipid metabolism and its impact on clinical strains of Mycobacterium tuberculosis.Microb Pathog. 2020 Sep;146:104250. doi: 10.1016/j.micpath.2020.104250. Epub 2020 May 11. Microb Pathog. 2020. PMID: 32407863 Review.
-
Host-pathogen coevolution in human tuberculosis.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):850-9. doi: 10.1098/rstb.2011.0316. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312052 Free PMC article. Review.
Cited by
-
Genotypes and drug resistance pattern of Mycobacterium tuberculosis complex among clinically diagnosed pulmonary tuberculosis patients.Front Public Health. 2024 Dec 2;12:1420685. doi: 10.3389/fpubh.2024.1420685. eCollection 2024. Front Public Health. 2024. PMID: 39687724 Free PMC article.
-
Transcriptional regulation and drug resistance in Mycobacterium tuberculosis.Front Cell Infect Microbiol. 2022 Sep 2;12:990312. doi: 10.3389/fcimb.2022.990312. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36118045 Free PMC article. Review.
-
Characterizing in vivo loss of virulence of an HN878 Mycobacterium tuberculosis isolate from a genetic duplication event.Tuberculosis (Edinb). 2022 Dec;137:102272. doi: 10.1016/j.tube.2022.102272. Epub 2022 Nov 2. Tuberculosis (Edinb). 2022. PMID: 36375278 Free PMC article.
-
Presence of CRISPR CAS-Like Sequences as a Proposed Mechanism for Horizontal Genetic Exchanges between Trichomonas vaginalis and Its Associated Virus: A Comparative Genomic Analysis with the First Report of a Putative CRISPR CAS Structures in Eukaryotic Cells.Biomed Res Int. 2023 Nov 27;2023:8069559. doi: 10.1155/2023/8069559. eCollection 2023. Biomed Res Int. 2023. PMID: 38058394 Free PMC article.
-
Mycobacterium tuberculosis complex molecular networks and their regulation: Implications of strain heterogeneity on epigenetic diversity and transcriptome regulation.Heliyon. 2023 Nov 19;9(12):e22611. doi: 10.1016/j.heliyon.2023.e22611. eCollection 2023 Dec. Heliyon. 2023. PMID: 38046135 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

