Degradation and Toxicity Analysis of a Reactive Textile Diazo Dye-Direct Red 81 by Newly Isolated Bacillus sp. DMS2
- PMID: 33072041
- PMCID: PMC7541843
- DOI: 10.3389/fmicb.2020.576680
Degradation and Toxicity Analysis of a Reactive Textile Diazo Dye-Direct Red 81 by Newly Isolated Bacillus sp. DMS2
Abstract
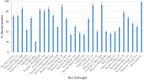
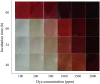
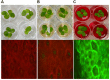
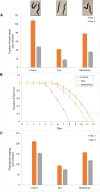
An efficient diazo dye degrading bacterial strain, Bacillus sp. DMS2 was isolated from a long-term textile dye polluted environment. The strain was assessed for its innate ability to completely degrade and detoxify Direct Red 81 (DR81) textile dye under microaerophilic conditions. The degradation ability of strain showed significant results on optimizing the nutritional and environmental parameters. Based on statistical models, maximum efficiency of decolorization achieved within 24 h for 100 mg/l of dye supplemented with glucose (0.02%), MgSO4 (0.002%) and urea (0.5%) at 30°C and pH (7.0). Moreover, a significant catabolic induction of a laccase and azoreductase suggested its vital role in degrading DR81 into three distinct metabolites (intermediates) as by-products. Further, toxicity analysis of intermediates were performed using seeds of common edible plants, aquatic plant (phytotoxicity) and the nematode model (animal toxicity), which confirmed the non-toxic nature of intermediates. Thus, the inclusive study of DMS2 showed promising efficiency in bioremediation approach for treating industrial effluents.
Keywords: Caenorhabditis elegans; Direct Red 81; Lemna minor; biodegradation; response surface methodology; toxicity.
Copyright © 2020 Amin, Rastogi, Chaubey, Jain, Divecha, Desai and Madamwar.
Figures


References
-
- Agrawal S., Tipre D., Patel B., Dave S. (2014). Optimization of triazo Acid Black 210 dye degradation by Providencia sp. SRS82 and elucidation of degradation pathway. Process Biochem. 49 110–119. 10.1016/j.procbio.2013.10.006 - DOI
-
- Akansha K., Chakraborty D., Sachan S. G. (2019). Decolorization and degradation of methyl orange by Bacillus stratosphericus SCA1007. Biocatalysis Agric. Biotechnol. 18:101044 10.1016/j.bcab.2019.101044 - DOI
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. A., Smith J. G., et al. (1997). Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons.
-
- Bedekar P. A., Kshirsagar S. D., Gholave A. R., Govindwar S. P. (2015). Degradation and detoxification of methylene blue dye adsorbed on water hyacinth in semi continuous anaerobic–aerobic bioreactors by novel microbial consortium-SB. RSC Adv. 5 99228–99239. 10.1039/C5RA17345K - DOI
LinkOut - more resources
Full Text Sources

