Forward Genetics Reveals a gatC-gatA Fusion Polypeptide Causes Mistranslation and Rifampicin Tolerance in Mycobacterium smegmatis
- PMID: 33072044
- PMCID: PMC7541841
- DOI: 10.3389/fmicb.2020.577756
Forward Genetics Reveals a gatC-gatA Fusion Polypeptide Causes Mistranslation and Rifampicin Tolerance in Mycobacterium smegmatis
Abstract
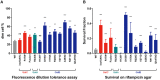
Most bacteria, including mycobacteria, utilize a two-step indirect tRNA aminoacylation pathway to generate correctly aminoacylated glutaminyl and asparaginyl tRNAs. This involves an initial step in which a non-discriminatory aminoacyl tRNA synthetase misacylates the tRNA, followed by a second step in which the essential amidotransferase, GatCAB, amidates the misacylated tRNA to its correct, cognate form. It had been previously demonstrated that mutations in gatA can mediate increased error rates specifically of glutamine to glutamate or asparagine to aspartate in protein synthesis. However, the role of mutations in gatB or gatC in mediating mistranslation are unknown. Here, we applied a forward genetic screen to enrich for mistranslating mutants of Mycobacterium smegmatis. The majority (57/67) of mutants had mutations in one of the gatCAB genes. Intriguingly, the most common mutation identified was an insertion in the 3' of gatC, abolishing its stop codon, and resulting in a fused GatC-GatA polypeptide. Modeling the effect of the fusion on GatCAB structure suggested a disruption of the interaction of GatB with the CCA-tail of the misacylated tRNA, suggesting a potential mechanism by which this mutation may mediate increased translational errors. Furthermore, we confirm that the majority of mutations in gatCAB that result in increased mistranslation also cause increased tolerance to rifampicin, although there was not a perfect correlation between mistranslation rates and degree of tolerance. Overall, our study identifies that mutations in all three gatCAB genes can mediate adaptive mistranslation and that mycobacteria are extremely tolerant to perturbation in the indirect tRNA aminoacylation pathway.
Keywords: GatCAB; Mycobacterium; amidotransferase; antibiotic tolerance; mistranslation; persisters.
Copyright © 2020 Cai, Su, Li and Javid.
Figures


References
-
- Curnow A. W., Hong K., Yuan R., Kim S., Martins O., Winkler W., et al. (1997). Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. U.S.A. 94 11819–11826. 10.1073/pnas.94.22.11819 - DOI - PMC - PubMed

