Role of BgaA as a Pneumococcal Virulence Factor Elucidated by Molecular Evolutionary Analysis
- PMID: 33072054
- PMCID: PMC7541833
- DOI: 10.3389/fmicb.2020.582437
Role of BgaA as a Pneumococcal Virulence Factor Elucidated by Molecular Evolutionary Analysis
Abstract
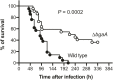
Streptococcus pneumoniae is a major cause of pneumonia, sepsis, and meningitis. Previously, we identified a novel virulence factor by investigating evolutionary selective pressure exerted on pneumococcal choline-binding cell surface proteins. Herein, we focus on another pneumococcal cell surface protein. Cell wall-anchoring proteins containing the LPXTG motif are conserved in Gram-positive bacteria. Our evolutionary analysis showed that among the examined genes, nanA and bgaA had high proportions of codon that were under significant negative selection. Both nanA and bgaA encode a multi-functional glycosidase that aids nutrient acquisition in a glucose-poor environment, pneumococcal adherence to host cells, and evasion from host immunity. However, several studies have shown that the role of BgaA is limited in a mouse pneumonia model, and it remains unclear if BgaA affects pneumococcal pathogenesis in a mouse sepsis model. To evaluate the distribution and pathogenicity of bgaA, we performed phylogenetic analysis and intravenous infection assay. In both Bayesian and maximum likelihood phylogenetic trees, the genetic distances between pneumococcal bgaA was small, and the cluster of pneumococcal bgaA did not contain other bacterial orthologs except for a Streptococcus gwangjuense gene. Evolutionary analysis and BgaA structure indicated BgaA active site was not allowed to change. The mouse infection assay showed that the deletion of bgaA significantly reduced host mortality. These results indicated that both nanA and bgaA encode evolutionally conserved pneumococcal virulence factors and that molecular evolutionary analysis could be a useful alternative strategy for identification of virulence factors.
Keywords: Streptococcus pneumoniae; bgaA; molecular evolutionary analysis; nanA; pneumococcal cell wall-anchoring proteins.
Copyright © 2020 Yamaguchi, Takemura, Higashi, Goto, Hirose, Sumitomo, Nakata, Uzawa and Kawabata.
Figures


Similar articles
-
Pneumococcal BgaA Promotes Host Organ Bleeding and Coagulation in a Mouse Sepsis Model.Front Cell Infect Microbiol. 2022 Jul 1;12:844000. doi: 10.3389/fcimb.2022.844000. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846740 Free PMC article.
-
BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells.Microbiology (Reading). 2011 Aug;157(Pt 8):2369-2381. doi: 10.1099/mic.0.045609-0. Epub 2011 May 20. Microbiology (Reading). 2011. PMID: 21602213 Free PMC article.
-
Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA.PLoS Pathog. 2014 Sep 11;10(9):e1004364. doi: 10.1371/journal.ppat.1004364. eCollection 2014 Sep. PLoS Pathog. 2014. PMID: 25210925 Free PMC article.
-
[Investigation of pneumococcal virulence factors in the infection process].Nihon Saikingaku Zasshi. 2020;75(2):173-183. doi: 10.3412/jsb.75.173. Nihon Saikingaku Zasshi. 2020. PMID: 33361653 Review. Japanese.
-
Pneumococcal virulence factors: structure and function.Microbiol Mol Biol Rev. 2001 Jun;65(2):187-207 ; first page, table of contents. doi: 10.1128/MMBR.65.2.187-207.2001. Microbiol Mol Biol Rev. 2001. PMID: 11381099 Free PMC article. Review.
Cited by
-
Epidemiological analysis of pneumococcal strains isolated at Yangon Children's Hospital in Myanmar via whole-genome sequencing-based methods.Microb Genom. 2021 Feb;7(2):000523. doi: 10.1099/mgen.0.000523. Microb Genom. 2021. PMID: 33565958 Free PMC article.
-
Pneumococcal BgaA Promotes Host Organ Bleeding and Coagulation in a Mouse Sepsis Model.Front Cell Infect Microbiol. 2022 Jul 1;12:844000. doi: 10.3389/fcimb.2022.844000. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846740 Free PMC article.
-
Role of Streptococcus pneumoniae extracellular glycosidases in immune evasion.Front Cell Infect Microbiol. 2023 Feb 3;13:1109449. doi: 10.3389/fcimb.2023.1109449. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36816580 Free PMC article. Review.
-
An innate pathogen sensing strategy involving ubiquitination of bacterial surface proteins.Sci Adv. 2023 Mar 22;9(12):eade1851. doi: 10.1126/sciadv.ade1851. Epub 2023 Mar 22. Sci Adv. 2023. PMID: 36947610 Free PMC article.
-
Editorial: Host-Pathogen Interactions During Pneumococcal Infection.Front Cell Infect Microbiol. 2021 Oct 25;11:752959. doi: 10.3389/fcimb.2021.752959. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34760720 Free PMC article. No abstract available.
References
-
- Beulin D. S. J., Radhakrishnan D., Suresh S. C., Sadasivan C., Yamaguchi M., Kawabata S., et al. (2017). Streptococcus pneumoniae surface protein PfbA is a versatile multidomain and multiligand-binding adhesin employing different binding mechanisms. FEBS J. 284 3404–3421. 10.1111/febs.14200 - DOI - PubMed
-
- Bricker A. L., Camilli A. (1999). Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172 131–135. - PubMed
LinkOut - more resources
Full Text Sources

