Towards a unified classification for human respiratory syncytial virus genotypes
- PMID: 33072402
- PMCID: PMC7552823
- DOI: 10.1093/ve/veaa052
Towards a unified classification for human respiratory syncytial virus genotypes
Abstract
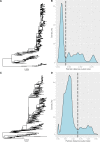
Since the first human respiratory syncytial virus (HRSV) genotype classification in 1998, inconsistent conclusions have been drawn regarding the criteria that define HRSV genotypes and their nomenclature, challenging data comparisons between research groups. In this study, we aim to unify the field of HRSV genotype classification by reviewing the different methods that have been used in the past to define HRSV genotypes and by proposing a new classification procedure, based on well-established phylogenetic methods. All available complete HRSV genomes (>12,000 bp) were downloaded from GenBank and divided into the two subgroups: HRSV-A and HRSV-B. From whole-genome alignments, the regions that correspond to the open reading frame of the glycoprotein G and the second hypervariable region (HVR2) of the ectodomain were extracted. In the resulting partial alignments, the phylogenetic signal within each fragment was assessed. Maximum likelihood phylogenetic trees were reconstructed using the complete genome alignments. Patristic distances were calculated between all pairs of tips in the phylogenetic tree and summarized as a density plot in order to determine a cutoff value at the lowest point following the major distance peak. Our data show that neither the HVR2 fragment nor the G gene contains sufficient phylogenetic signal to perform reliable phylogenetic reconstruction. Therefore, whole-genome alignments were used to determine HRSV genotypes. We define a genotype using the following criteria: a bootstrap support of 70 per cent for the respective clade and a maximum patristic distance between all members of the clade of ≤0.018 substitutions per site for HRSV-A or ≤0.026 substitutions per site for HRSV-B. By applying this definition, we distinguish twenty-three genotypes within subtype HRSV-A and six genotypes within subtype HRSV-B. Applying the genotype criteria on subsampled data sets confirmed the robustness of the method.
Keywords: classification; genotypes; human respiratory syncytial virus.
© The Author(s) 2020. Published by Oxford University Press.
Figures



References
-
- Anderson L. J. et al. (1985) ‘Antigenic Characterization of Respiratory Syncytial Virus Strains with Monoclonal Antibodies’, The Journal of Infectious Diseases, 151: 626–33. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

