Characterization of Pneumococcal Colonization Dynamics and Antimicrobial Resistance Using Shotgun Metagenomic Sequencing in Intensively Sampled South African Infants
- PMID: 33072693
- PMCID: PMC7536305
- DOI: 10.3389/fpubh.2020.543898
Characterization of Pneumococcal Colonization Dynamics and Antimicrobial Resistance Using Shotgun Metagenomic Sequencing in Intensively Sampled South African Infants
Abstract
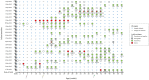
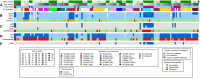
Background: There remains a significant proportion of deaths due to pneumococcal pneumonia in infants from low- and middle-income countries despite the marginal global declines recorded in the past decade. Monitoring changes in pneumococcal carriage is key to understanding vaccination-induced shifts in the ecology of carriage, patterns of antimicrobial resistance, and impact on health. We longitudinally investigated pneumococcal carriage dynamics in PCV-13 vaccinated infants by collecting nasopharyngeal (NP) samples at 2-weekly intervals from birth through the first year of life from 137 infants. As a proof of concept, 196 NP samples were retrieved from a subset of 23 infants to explore strain-level pneumococcal colonization patterns and associated antimicrobial-resistance determinants. These were selected on the basis of changes in serotype and antibiogram over time. NP samples underwent short-term enrichment for streptococci prior to total nucleic acid extraction and whole metagenome shotgun sequencing (WMGS). Reads were assembled and aligned to pneumococcal reference genomes for the extraction of pneumococcal and non-pneumococcal bacterial reads. Pneumococcal contigs were aligned to the Antibiotic Resistance Gene-ANNOTation database of acquired AMR genes. In silico pneumococcal capsular and multilocus sequence typing were performed. Results: Of the 196 samples sequenced, 174 had corresponding positive cultures for pneumococci, of which, 152 were assigned an in silico serotype. Metagenomic sequencing detected a single pneumococcal serotype in 85% (129/152), and co-colonization in 15% (23/152) of the samples. Twenty-two different pneumococcal serotypes were identified, with 15B/15C and 16F being the most common non-PCV13 serotypes, while 23F and 19A were the most common PCV13 serotypes. Twenty-six different sequence types (STs), including four novel STs were identified in silico. Mutations in the folA and folP genes, associated with cotrimoxazole resistance, were detected in 89% (87/98) of cotrimoxazole-non-susceptible pneumococci, as well as in the pbp1a and pbp2x genes, in penicillin non-susceptible ST705215B/15C isolates. Conclusions: Metagenomic sequencing of NP samples is a valuable culture-independent technique for a detailed evaluation of the pneumococcal component and resistome of the NP microbiome. This method allowed for the detection of novel STs, as well as co-colonization, with a predominance of non-PCV13 serotypes in this cohort. Forty-eight resistance genes, as well as mutations associated with resistance were detected, but the correlation with phenotypic non-susceptibility was lower than expected.
Keywords: Streptococcus pneumoniae; multi locus sequence typing; nasopharyngeal; pneumococcal conjugate vaccine; resistance determinants; serotypes; shotgun metagenomic sequencing.
Copyright © 2020 Manenzhe, Dube, Wright, Lennard, Mounaud, Lo, Zar, Nierman, Nicol and Moodley.
Figures

Similar articles
-
Longitudinal changes in the nasopharyngeal resistome of South African infants using shotgun metagenomic sequencing.PLoS One. 2020 Apr 22;15(4):e0231887. doi: 10.1371/journal.pone.0231887. eCollection 2020. PLoS One. 2020. PMID: 32320455 Free PMC article.
-
Nasopharyngeal Carriage of Antimicrobial-Resistant Pneumococci in an Intensively Sampled South African Birth Cohort.Front Microbiol. 2019 Mar 27;10:610. doi: 10.3389/fmicb.2019.00610. eCollection 2019. Front Microbiol. 2019. PMID: 30972052 Free PMC article.
-
Strain Level Streptococcus Colonization Patterns during the First Year of Life.Front Microbiol. 2017 Sep 6;8:1661. doi: 10.3389/fmicb.2017.01661. eCollection 2017. Front Microbiol. 2017. PMID: 28932211 Free PMC article.
-
Impact of pneumococcal conjugate vaccine on infections caused by antibiotic-resistant Streptococcus pneumoniae.Clin Microbiol Infect. 2009 Apr;15 Suppl 3:16-20. doi: 10.1111/j.1469-0691.2009.02726.x. Clin Microbiol Infect. 2009. PMID: 19366365 Review.
-
State-of-the-art in the pneumococcal field: Proceedings of the 11th International Symposium on Pneumococci and Pneumococcal Diseases (ISPPD-11).Pneumonia (Nathan). 2020 Feb 5;12:2. doi: 10.1186/s41479-019-0064-y. eCollection 2020. Pneumonia (Nathan). 2020. PMID: 32042572 Free PMC article. Review.
Cited by
-
Strain-level resolution and pneumococcal carriage dynamics by single-molecule real-time (SMRT) sequencing of the plyNCR marker: a longitudinal study in Swiss infants.Microbiome. 2022 Sep 22;10(1):152. doi: 10.1186/s40168-022-01344-6. Microbiome. 2022. PMID: 36138483 Free PMC article.
-
Expansion of pneumococcal serotype 23F and 14 lineages with genotypic changes in capsule polysaccharide locus and virulence gene profiles post introduction of pneumococcal conjugate vaccine in Blantyre, Malawi.Microb Genom. 2024 Jun;10(6):001264. doi: 10.1099/mgen.0.001264. Microb Genom. 2024. PMID: 38896467 Free PMC article.
-
Streptococcus pneumoniae epidemiology, pathogenesis and control.Nat Rev Microbiol. 2025 Apr;23(4):256-271. doi: 10.1038/s41579-024-01116-z. Epub 2024 Nov 6. Nat Rev Microbiol. 2025. PMID: 39506137 Review.
References
-
- GBD 2015 Mortality and Causes of Death Collaborators . Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the global burden of disease study 2015. Lancet. (2016) 388:1459–544. 10.1016/S0140-6736(16)31012-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

