Proteome analysis of Saccharomyces cerevisiae after methyl methane sulfonate (MMS) treatment
- PMID: 33072891
- PMCID: PMC7548944
- DOI: 10.1016/j.bbrep.2020.100820
Proteome analysis of Saccharomyces cerevisiae after methyl methane sulfonate (MMS) treatment
Abstract
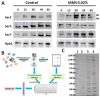
The treatment of methyl methane sulfonate (MMS) increases sensitivity to the DNA damage which, further leads to the cell death followed by a cell cycle delay. Delay in the cell cycle is because of the change in global transcription regulation which results into proteome change. There are several microarray studies on the transcriptome changes after MMS treatment, but very few studies are reported related to proteome change. The proteome analysis in this report identified subgroups of proteins, belonging to known cell cycle regulators, metabolic pathways and protein folding. About 53 proteins were identified by MS/MS and found that 36 of them were induced, 10 were repressed and few of them showed insignificant change. Our results indicated the change in the interactome as well as phosphorylation status of carboxy terminal domain (CTD) of RNA Polymerase II (RNAP-II) after MMS treatment. The RNAP-II complex was affinity purified and ~1640 peptides were identified using nano LC/MS corresponding to 27 interacting proteins along with the twelve RNAP-II subunit. These identified proteins participated in the repair of the damage, changes the function of the main energetic pathways and the carbon flux in various end products. The main metabolic enzymes in the glycolysis, pyruvate phosphate and amino acid biosynthesis pathways showed significant change. Our results indicate that DNA damage is somehow related to these pathways and is co-regulated simultaneously.
Keywords: DNA damage; Mass spectrometry (MS); Methyl methane sulfonate (MMS); Proteomics; RNA polymerase II-CTD.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
DNA Replication Stress Phosphoproteome Profiles Reveal Novel Functional Phosphorylation Sites on Xrs2 in Saccharomyces cerevisiae.Genetics. 2016 May;203(1):353-68. doi: 10.1534/genetics.115.185231. Epub 2016 Mar 26. Genetics. 2016. PMID: 27017623 Free PMC article.
-
Cell-cycle and DNA damage regulation of the DNA mismatch repair protein Msh2 occurs at the transcriptional and post-transcriptional level.DNA Repair (Amst). 2013 Feb 1;12(2):97-109. doi: 10.1016/j.dnarep.2012.11.002. Epub 2012 Dec 20. DNA Repair (Amst). 2013. PMID: 23261051 Free PMC article.
-
Proteomics studies of the interactome of RNA polymerase II C-terminal repeated domain.BMC Res Notes. 2015 Oct 29;8:616. doi: 10.1186/s13104-015-1569-y. BMC Res Notes. 2015. PMID: 26515650 Free PMC article.
-
Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control.J Cell Physiol. 2002 Feb;190(2):160-9. doi: 10.1002/jcp.10058. J Cell Physiol. 2002. PMID: 11807820 Review.
-
The Old and New Testaments of gene regulation. Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD.Transcription. 2014;5(3):e28674. doi: 10.4161/trns.28674. Transcription. 2014. PMID: 25764332 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

