Identification of novel mutations in the methyltransferase complex (Nsp10-Nsp16) of SARS-CoV-2
- PMID: 33072893
- PMCID: PMC7547569
- DOI: 10.1016/j.bbrep.2020.100833
Identification of novel mutations in the methyltransferase complex (Nsp10-Nsp16) of SARS-CoV-2
Abstract
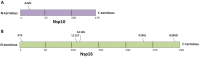
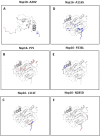
A recent outburst of the pandemic caused by a member of the coronaviridae family identified as SARS-CoV-2. The highly contagious nature of the virus allows it to spread rapidly worldwide and caused severe healthcare and economic distress. So far, no proper line of treatment or vaccines has been available against SARS-CoV-2. Since, the infected people rapidly increased, causing the saturation of healthcare systems with coronavirus disease (COVID-19) patients. As the virus spread to new locations it also acquired various mutations. Here, in this study, we focused on identifying mutations in one of the crucial complex of SARS-CoV-2, the Nsp10-Nsp16 2'-O-methyltransferase complex. This complex plays indispensable role in the post-transcriptional modifications of viral RNA by its capping. We analysed 208 sequences of Nsp10-Nsp16 reported from India and compared with first reported sequence from Wuhan, China. Our analysis revealed a single mutation in Nsp10 and five mutations in Nsp16 protein. We also show that these mutations are leading to alteration in the secondary structure of Nsp10-Nsp16. Further, the protein modelling studies revealed that the mutation of both Nsp10-Nsp16 impacts the protein dynamicity and stability. Altogether, this study provides novel insights into the variations observed in the proteins of SARS-CoV-2 that might have functional consequences.
Keywords: COVID-19; India; Infectious diseases; Mutations; Nsp10-Nsp16; SARS-CoV-2.
© 2020 The Author.
Conflict of interest statement
Authors declare no conflict of interests.
Figures


References
-
- Zhou P., Yang X. Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R. Di, Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

