SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations
- PMID: 33073312
- PMCID: PMC7568947
- DOI: 10.1007/s00216-020-02958-1
SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations
Abstract
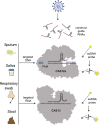
The unprecedented global pandemic known as SARS-CoV-2 has exercised to its limits nearly all aspects of modern viral diagnostics. In doing so, it has illuminated both the advantages and limitations of current technologies. Tremendous effort has been put forth to expand our capacity to diagnose this deadly virus. In this work, we put forth key observations in the functionality of current methods for SARS-CoV-2 diagnostic testing. These methods include nucleic acid amplification-, CRISPR-, sequencing-, antigen-, and antibody-based detection methods. Additionally, we include analysis of equally critical aspects of COVID-19 diagnostics, including sample collection and preparation, testing models, and commercial response. We emphasize the integrated nature of assays, wherein issues in sample collection and preparation could impact the overall performance in a clinical setting.
Keywords: COVID-19; CRISPR; RT-PCR; SARS-CoV-2; Sequencing; Serological testing.
Conflict of interest statement
Harikrishnan Jayamohan is a former employee of Roche Sequencing Solutions Inc. and owns shares in Roche Holding AG. All other authors have no conflicts to declare.
Figures



References
-
- John Hopkins Center for Health Security, Comparison of national RT-PCR primers, probes, and protocols for SARS-CoV-2 diagnostics, Centerforhealthsecurity.Org. 2020;5. https://www.centerforhealthsecurity.org/resources/COVID-19/COVID-19-fact....
-
- Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020:0–28. 10.1021/acsnano.0c02624. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

