Uncovering ecological state dynamics with hidden Markov models
- PMID: 33073921
- PMCID: PMC7702077
- DOI: 10.1111/ele.13610
Uncovering ecological state dynamics with hidden Markov models
Erratum in
-
Corrigendum.Ecol Lett. 2021 May;24(5):1117. doi: 10.1111/ele.13709. Epub 2021 Mar 16. Ecol Lett. 2021. PMID: 33724634 Free PMC article. No abstract available.
Abstract
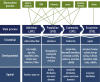
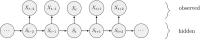
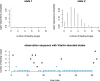
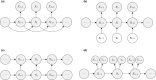
Ecological systems can often be characterised by changes among a finite set of underlying states pertaining to individuals, populations, communities or entire ecosystems through time. Owing to the inherent difficulty of empirical field studies, ecological state dynamics operating at any level of this hierarchy can often be unobservable or 'hidden'. Ecologists must therefore often contend with incomplete or indirect observations that are somehow related to these underlying processes. By formally disentangling state and observation processes based on simple yet powerful mathematical properties that can be used to describe many ecological phenomena, hidden Markov models (HMMs) can facilitate inferences about complex system state dynamics that might otherwise be intractable. However, HMMs have only recently begun to gain traction within the broader ecological community. We provide a gentle introduction to HMMs, establish some common terminology, review the immense scope of HMMs for applied ecological research and provide a tutorial on implementation and interpretation. By illustrating how practitioners can use a simple conceptual template to customise HMMs for their specific systems of interest, revealing methodological links between existing applications, and highlighting some practical considerations and limitations of these approaches, our goal is to help establish HMMs as a fundamental inferential tool for ecologists.
Keywords: Behavioural ecology; community ecology; ecosystem ecology; hierarchical model; movement ecology; observation error; population ecology; state-space model; time series.
Published 2020. This article is a U.S. Government work and is in the public domain in the USA. Ecology Letters published by John Wiley & Sons Ltd.
Figures
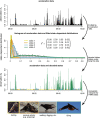
Similar articles
-
Classifying movement behaviour in relation to environmental conditions using hidden Markov models.J Anim Ecol. 2009 Nov;78(6):1113-23. doi: 10.1111/j.1365-2656.2009.01583.x. Epub 2009 Jun 26. J Anim Ecol. 2009. PMID: 19563470
-
Objective classification of latent behavioral states in bio-logging data using multivariate-normal hidden Markov models.Ecol Appl. 2015 Jul;25(5):1244-58. doi: 10.1890/14-0862.1. Ecol Appl. 2015. PMID: 26485953
-
Flexible and practical modeling of animal telemetry data: hidden Markov models and extensions.Ecology. 2012 Nov;93(11):2336-42. doi: 10.1890/11-2241.1. Ecology. 2012. PMID: 23236905
-
Successional theories.Biol Rev Camb Philos Soc. 2023 Dec;98(6):2049-2077. doi: 10.1111/brv.12995. Epub 2023 Jul 16. Biol Rev Camb Philos Soc. 2023. PMID: 37455023 Review.
-
Accounting for uncertainty in ecological analysis: the strengths and limitations of hierarchical statistical modeling.Ecol Appl. 2009 Apr;19(3):553-70. doi: 10.1890/07-0744.1. Ecol Appl. 2009. PMID: 19425416 Review.
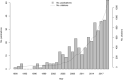
Cited by
-
Fall migration, oceanic movement, and site residency patterns of eastern red bats (Lasiurus borealis) on the mid-Atlantic Coast.Mov Ecol. 2023 Jun 14;11(1):35. doi: 10.1186/s40462-023-00398-x. Mov Ecol. 2023. PMID: 37316899 Free PMC article.
-
An introduction to statistical models used to characterize species-habitat associations with animal movement data.Mov Ecol. 2025 Apr 17;13(1):27. doi: 10.1186/s40462-025-00549-2. Mov Ecol. 2025. PMID: 40247418 Free PMC article. Review.
-
Unlocking sea turtle diving behaviour from low-temporal resolution time-depth recorders.Sci Rep. 2025 Jun 6;15(1):19934. doi: 10.1038/s41598-025-05336-y. Sci Rep. 2025. PMID: 40481176 Free PMC article.
-
Corrigendum.Ecol Lett. 2021 May;24(5):1117. doi: 10.1111/ele.13709. Epub 2021 Mar 16. Ecol Lett. 2021. PMID: 33724634 Free PMC article. No abstract available.
-
Incorporating sparse labels into hidden Markov models using weighted likelihoods improves accuracy and interpretability in biologging studies.PLoS One. 2025 Jun 18;20(6):e0325321. doi: 10.1371/journal.pone.0325321. eCollection 2025. PLoS One. 2025. PMID: 40531913 Free PMC article.
References
-
- Abercrombie, S.P. & Friedl, M.A. (2015). Improving the consistency of multitemporal land cover maps using a hidden Markov model. IEEE Trans. Geosci. Remote Sens., 54, 703–713.
-
- Adam, T. , Griffiths, C.A. , Leos‐Barajas, V. , Meese, E.N. , Lowe, C.G. , Blackwell, P.G. et al (2019a). Joint modelling of multi‐scale animal movement data using hierarchical hidden Markov models. Methods Ecol. Evol., 10, 1536–1550.
-
- Adam, T. , Langrock, R. & Weiß, C.H. (2019b). Penalized estimation of flexible hidden Markov models for time series of counts. METRON, 77, 87–104.
-
- Altman, R.M. (2007). Mixed hidden Markov models: an extension of the hidden Markov model to the longitudinal data setting. Journal of the American Statistical Association, 102, 201–210.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

