Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells
- PMID: 33075123
- PMCID: PMC7579740
- DOI: 10.1084/jem.20200891
Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells
Abstract
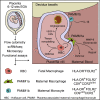
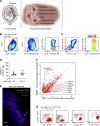
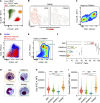
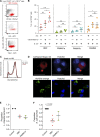
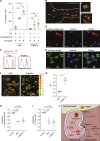
Hofbauer cells (HBCs) are a population of macrophages found in high abundance within the stroma of the first-trimester human placenta. HBCs are the only fetal immune cell population within the stroma of healthy placenta. However, the functional properties of these cells are poorly described. Aligning with their predicted origin via primitive hematopoiesis, we find that HBCs are transcriptionally similar to yolk sac macrophages. Phenotypically, HBCs can be identified as HLA-DR-FOLR2+ macrophages. We identify a number of factors that HBCs secrete (including OPN and MMP-9) that could affect placental angiogenesis and remodeling. We determine that HBCs have the capacity to play a defensive role, where they are responsive to Toll-like receptor stimulation and are microbicidal. Finally, we also identify a population of placenta-associated maternal macrophages (PAMM1a) that adhere to the placental surface and express factors, such as fibronectin, that may aid in repair.
© 2020 Thomas et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






Comment in
-
Gatekeepers of the fetus: Characterization of placental macrophages.J Exp Med. 2021 Jan 4;218(1):e20202071. doi: 10.1084/jem.20202071. J Exp Med. 2021. PMID: 33601417 Free PMC article.
References
-
- Boyd J.D. and Hamilton W.J.. 1970. Stroma of villi. In The Human Placenta. Heffer, Cambridge.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

