Long genes are more frequently affected by somatic mutations and show reduced expression in Alzheimer's disease: Implications for disease etiology
- PMID: 33075204
- PMCID: PMC8048495
- DOI: 10.1002/alz.12211
Long genes are more frequently affected by somatic mutations and show reduced expression in Alzheimer's disease: Implications for disease etiology
Abstract
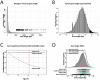
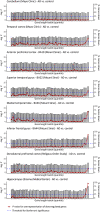
Aging, the greatest risk factor for Alzheimer's disease (AD), may lead to the accumulation of somatic mutations in neurons. We investigated whether somatic mutations, specifically in longer genes, are implicated in AD etiology. First, we modeled the theoretical likelihood of genes being affected by aging-induced somatic mutations, dependent on their length. We then tested this model and found that long genes are indeed more affected by somatic mutations and that their expression is more frequently reduced in AD brains. Furthermore, using gene-set enrichment analysis, we investigated the potential consequences of such long gene disruption. We found that long genes are involved in synaptic adhesion and other synaptic pathways that are predicted to be inhibited in the brains of AD patients. Taken together, our findings indicate that long gene-dependent synaptic impairment may contribute to AD pathogenesis.
Keywords: Alzheimer's disease; DNA damage; long genes; somatic mutations; synaptic adhesion.
© 2020 The Authors. Alzheimer's & Dementia published by Wiley Periodicals, Inc. on behalf of Alzheimer's Association.
Conflict of interest statement
Geert Poelmans is director of DrugTarget ID, Ltd. (The Netherlands). All other authors declare no conflicts of interest.
Figures

References
-
- Patterson C. World Alzheimer Report 2018: The State of the Art of Dementia Research: New Frontiers. London, UK: Alzheimer's Disease International (ADI); 2018.
-
- van der Lee SJ, Wolters FJ, Ikram MK, et al. The effect of APOE and other common genetic variants on the onset of Alzheimer's disease and dementia: a community‐based cohort study. Lancet Neurol. 2018;17(5):434‐444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AG046152/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- R01 AG030146/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

