S100A11 Promotes Liver Steatosis via FOXO1-Mediated Autophagy and Lipogenesis
- PMID: 33075563
- PMCID: PMC7841444
- DOI: 10.1016/j.jcmgh.2020.10.006
S100A11 Promotes Liver Steatosis via FOXO1-Mediated Autophagy and Lipogenesis
Abstract
Background & aims: Nonalcoholic fatty liver disease (NAFLD) is becoming a severe liver disorder worldwide. Autophagy plays a critical role in liver steatosis. However, the role of autophagy in NAFLD remains exclusive and under debate. In this study, we investigated the role of S100 calcium binding protein A11 (S100A11) in the pathogenesis of hepatic steatosis.
Methods: We performed liver proteomics in a well-established tree shrew model of NAFLD. The expression of S100A11 in different models of NAFLD was detected by Western blot and/or quantitative polymerase chain reaction. Liver S100A11 overexpression mice were generated by injecting a recombinant adenovirus gene transfer vector through the tail vein and then induced by a high-fat and high-cholesterol diet. Cell lines with S100a11 stable overexpression were established with a recombinant lentiviral vector. The lipid content was measured with either Bodipy staining, Oil Red O staining, gas chromatography, or a triglyceride kit. The autophagy and lipogenesis were detected in vitro and in vivo by Western blot and quantitative polymerase chain reaction. The functions of Sirtuin 1, histone deacetylase 6 (HDAC6), and FOXO1 were inhibited by specific inhibitors. The interactions between related proteins were analyzed by a co-immunoprecipitation assay and immunofluorescence analysis.
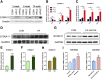
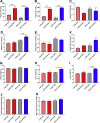
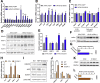
Results: The expression of S100A11 was up-regulated significantly in a time-dependent manner in the tree shrew model of NAFLD. S100A11 expression was induced consistently in oleic acid-treated liver cells as well as the livers of mice fed a high-fat diet and NAFLD patients. Both in vitro and in vivo overexpression of S100A11 could induce hepatic lipid accumulation. Mechanistically, overexpression of S100A11 activated an autophagy and lipogenesis process through up-regulation and acetylation of the transcriptional factor FOXO1, consequently promoting lipogenesis and lipid accumulation in vitro and in vivo. Inhibition of HDAC6, a deacetylase of FOXO1, showed similar phenotypes to S100A11 overexpression in Hepa 1-6 cells. S100A11 interacted with HDAC6 to inhibit its activity, leading to the release and activation of FOXO1. Under S100A11 overexpression, the inhibition of FOXO1 and autophagy could alleviate the activated autophagy as well as up-regulated lipogenic genes. Both FOXO1 and autophagy inhibition and Dgat2 deletion could reduce liver cell lipid accumulation significantly.
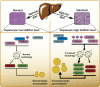
Conclusions: A high-fat diet promotes liver S100A11 expression, which may interact with HDAC6 to block its binding to FOXO1, releasing or increasing the acetylation of FOXO1, thus activating autophagy and lipogenesis, and accelerating lipid accumulation and liver steatosis. These findings indicate a completely novel S100A11-HDAC6-FOXO1 axis in the regulation of autophagy and liver steatosis, providing potential possibilities for the treatment of NAFLD.
Keywords: Autophagy; FOXO1; Lipid Metabolism; NAFLD; S100A11.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures















Comment in
-
S100A11 Overexpression Promotes Fatty Liver Diseases via Increased Autophagy?Cell Mol Gastroenterol Hepatol. 2021;11(3):885-886. doi: 10.1016/j.jcmgh.2020.11.013. Epub 2020 Dec 5. Cell Mol Gastroenterol Hepatol. 2021. PMID: 33290740 Free PMC article. No abstract available.
References
-
- Khan R., Bril F., Cusi K., Newsome P.N. Modulation of insulin resistance in NAFLD. Hepatology. 2019;70:711–724. - PubMed
-
- Younossi Z.M. Non-alcoholic fatty liver disease – a global public health perspective. J Hepatol. 2019;70:531–544. - PubMed
-
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Loomba R., Sanyal A.J. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

