A Cell-Based Capture Assay for Rapid Virus Detection
- PMID: 33076296
- PMCID: PMC7602404
- DOI: 10.3390/v12101165
A Cell-Based Capture Assay for Rapid Virus Detection
Abstract
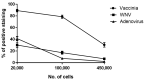
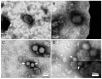
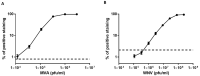
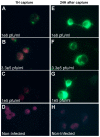
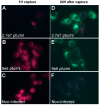
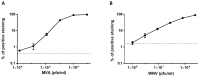
Routine methods for virus detection in clinical specimens rely on a variety of sensitive methods, such as genetic, cell culture and immuno-based assays. It is imperative that the detection assays would be reliable, reproducible, sensitive and rapid. Isolation of viruses from clinical samples is crucial for deeper virus identification and analysis. Here we introduce a rapid cell-based assay for isolation and detection of viruses. As a proof of concept several model viruses including West Nile Virus (WNV), Modified Vaccinia Ankara (MVA) and Adenovirus were chosen. Suspended Vero cells were employed to capture the viruses following specific antibody labeling which enables their detection by flow cytometry and immuno-fluorescence microscopy assays. Using flow cytometry, a dose response analysis was performed in which 3.6e4 pfu/mL and 1e6 pfu/mL of MVA and WNV could be detected within two hours, respectively. When spiked to commercial pooled human serum, detection sensitivity was slightly reduced to 3e6 pfu/mL for WNV, but remained essentially the same for MVA. In conclusion, the study demonstrates a robust and rapid methodology for virus detection using flow cytometry and fluorescence microscopy. We propose that this proof of concept may prove useful in identifying future pathogens.
Keywords: Adenovirus; Modified Vaccinia Ankara; West Nile Virus; flow cytometry; immuno-fluorescence assay; transmission electron microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

