Implementation of homology based and non-homology based computational methods for the identification and annotation of orphan enzymes: using Mycobacterium tuberculosis H37Rv as a case study
- PMID: 33076816
- PMCID: PMC7574302
- DOI: 10.1186/s12859-020-03794-x
Implementation of homology based and non-homology based computational methods for the identification and annotation of orphan enzymes: using Mycobacterium tuberculosis H37Rv as a case study
Abstract
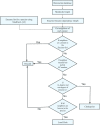
Background: Homology based methods are one of the most important and widely used approaches for functional annotation of high-throughput microbial genome data. A major limitation of these methods is the absence of well-characterized sequences for certain functions. The non-homology methods based on the context and the interactions of a protein are very useful for identifying missing metabolic activities and functional annotation in the absence of significant sequence similarity. In the current work, we employ both homology and context-based methods, incrementally, to identify local holes and chokepoints, whose presence in the Mycobacterium tuberculosis genome is indicated based on its interaction with known proteins in a metabolic network context, but have not been annotated. We have developed two computational procedures using network theory to identify orphan enzymes ('Hole finding protocol') coupled with the identification of candidate proteins for the predicted orphan enzyme ('Hole filling protocol'). We propose an integrated interaction score based on scores from the STRING database to identify candidate protein sequences for the orphan enzymes from M. tuberculosis, as a case study, which are most likely to perform the missing function.
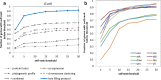
Results: The application of an automated homology-based enzyme identification protocol, ModEnzA, on M. tuberculosis genome yielded 56 novel enzyme predictions. We further predicted 74 putative local holes, 6 choke points, and 3 high confidence local holes in the genome using 'Hole finding protocol'. The 'Hole-filling protocol' was validated on the E. coli genome using artificial in-silico enzyme knockouts where our method showed 25% increased accuracy, compared to other methods, in assigning the correct sequence for the knocked-out enzyme amongst the top 10 ranks. The method was further validated on 8 additional genomes.
Conclusions: We have developed methods that can be generalized to augment homology-based annotation to identify missing enzyme coding genes and to predict a candidate protein for them. For pathogens such as M. tuberculosis, this work holds significance in terms of increasing the protein repertoire and thereby, the potential for identifying novel drug targets.
Keywords: Chokepoints; Genome context-based annotation; Global hole; Homology based method; Local hole; Missing enzyme; ModEnzA; Non-homology based methods.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases.BMC Bioinformatics. 2004 Jun 9;5:76. doi: 10.1186/1471-2105-5-76. BMC Bioinformatics. 2004. PMID: 15189570 Free PMC article.
-
Functional annotation of putative aminoglycoside antibiotic modifying proteins in Mycobacterium tuberculosis H37Rv.J Antibiot (Tokyo). 2003 Feb;56(2):135-42. doi: 10.7164/antibiotics.56.135. J Antibiot (Tokyo). 2003. PMID: 12715873
-
Accurate prediction of protein functional class from sequence in the Mycobacterium tuberculosis and Escherichia coli genomes using data mining.Yeast. 2000 Dec;17(4):283-93. doi: 10.1002/1097-0061(200012)17:4<283::AID-YEA52>3.0.CO;2-F. Yeast. 2000. PMID: 11119305 Free PMC article.
-
Functional assignment of Mycobacterium tuberculosis proteome revealed by genome-scale fold-recognition.Tuberculosis (Edinb). 2013 Jan;93(1):40-6. doi: 10.1016/j.tube.2012.11.008. Epub 2013 Jan 1. Tuberculosis (Edinb). 2013. PMID: 23287603 Review.
-
Using comparative genome analysis to identify problems in annotated microbial genomes.Microbiology (Reading). 2010 Jul;156(Pt 7):1909-1917. doi: 10.1099/mic.0.033811-0. Epub 2010 Apr 29. Microbiology (Reading). 2010. PMID: 20430813 Review.
Cited by
-
Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants.Pharmaceuticals (Basel). 2022 Sep 12;15(9):1136. doi: 10.3390/ph15091136. Pharmaceuticals (Basel). 2022. PMID: 36145357 Free PMC article.
-
Functional prediction of proteins from the human gut archaeome.ISME Commun. 2024 Jan 10;4(1):ycad014. doi: 10.1093/ismeco/ycad014. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38486809 Free PMC article.
-
Identification of a novel gene required for competitive growth at high temperature in the thermotolerant yeast Kluyveromyces marxianus.Microbiology (Reading). 2022 Mar;168(3):001148. doi: 10.1099/mic.0.001148. Microbiology (Reading). 2022. PMID: 35333706 Free PMC article.
-
An informatic workflow for the enhanced annotation of excretory/secretory proteins of Haemonchus contortus.Comput Struct Biotechnol J. 2023 Mar 18;21:2696-2704. doi: 10.1016/j.csbj.2023.03.025. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37143762 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

