Macrophage-Associated Lipin-1 Promotes β-Oxidation in Response to Proresolving Stimuli
- PMID: 33077427
- PMCID: PMC7739271
- DOI: 10.4049/immunohorizons.2000047
Macrophage-Associated Lipin-1 Promotes β-Oxidation in Response to Proresolving Stimuli
Abstract
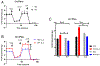
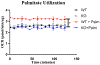
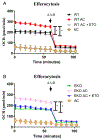
Macrophages reprogram their metabolism to promote appropriate responses. Proresolving macrophages primarily use fatty acid oxidation as an energy source. Metabolites generated during the catabolism of fatty acids aid in the resolution of inflammation and tissue repair, but the regulatory mechanisms that control lipid metabolism in macrophages are not fully elucidated. Lipin-1, a phosphatidic acid phosphatase that has transcriptional coregulator activity, regulates lipid metabolism in a variety of cells. In this current study, we show that lipin-1 is required for increased oxidative phosphorylation in IL-4 stimulated mouse (Mus musculus) macrophages. We also show that the transcriptional coregulatory function of lipin-1 is required for β-oxidation in response to palmitate (free fatty acid) and apoptotic cell (human) stimulation. Mouse bone marrow-derived macrophages lacking lipin-1 have a reduction in critical TCA cycle metabolites following IL-4 stimulation, suggesting a break in the TCA cycle that is supportive of lipid synthesis rather than lipid catabolism. Together, our data demonstrate that lipin-1 regulates cellular metabolism in macrophages in response to proresolving stimuli and highlights the importance of aligning macrophage metabolism with macrophage phenotype.
Copyright © 2020 The Authors.
Conflict of interest statement
DISCLOSURES
The authors have no financial conflicts of interest.
Figures

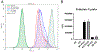


Similar articles
-
Lipin-1 restrains macrophage lipid synthesis to promote inflammation resolution.J Immunol. 2025 Jan 1;214(1):85-103. doi: 10.1093/jimmun/vkae010. J Immunol. 2025. PMID: 40073265
-
Lipin-1 Contributes to IL-4 Mediated Macrophage Polarization.Front Immunol. 2020 May 5;11:787. doi: 10.3389/fimmu.2020.00787. eCollection 2020. Front Immunol. 2020. PMID: 32431707 Free PMC article.
-
Leukemia inhibitory factor drives transcriptional programs that promote lipid accumulation and M2 polarization in macrophages.J Leukoc Biol. 2024 Dec 31;117(1):qiae178. doi: 10.1093/jleuko/qiae178. J Leukoc Biol. 2024. PMID: 39178293
-
The role of lipin-1 in the pathogenesis of alcoholic fatty liver.Alcohol Alcohol. 2015 Mar;50(2):146-51. doi: 10.1093/alcalc/agu102. Epub 2015 Jan 16. Alcohol Alcohol. 2015. PMID: 25595739
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
Cited by
-
Role of lipins in cardiovascular diseases.Lipids Health Dis. 2023 Nov 14;22(1):196. doi: 10.1186/s12944-023-01961-6. Lipids Health Dis. 2023. PMID: 37964368 Free PMC article. Review.
-
Myeloid-associated lipin-1 transcriptional co-regulatory activity is atheroprotective.Atherosclerosis. 2021 Aug;330:76-84. doi: 10.1016/j.atherosclerosis.2021.06.927. Epub 2021 Jul 1. Atherosclerosis. 2021. PMID: 34256308 Free PMC article.
-
Lipin-1 restrains macrophage lipid synthesis to promote inflammation resolution.bioRxiv [Preprint]. 2023 Oct 25:2023.10.23.563587. doi: 10.1101/2023.10.23.563587. bioRxiv. 2023. Update in: J Immunol. 2025 Jan 01;214(1):85-103. doi: 10.1093/jimmun/vkae010. PMID: 37961352 Free PMC article. Updated. Preprint.
-
Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer.Int J Mol Sci. 2021 Apr 23;22(9):4419. doi: 10.3390/ijms22094419. Int J Mol Sci. 2021. PMID: 33922580 Free PMC article. Review.
-
Metabolic Consequences of Efferocytosis and its Impact on Atherosclerosis.Immunometabolism. 2021;3(2):e210017. doi: 10.20900/immunometab20210017. Epub 2021 Mar 31. Immunometabolism. 2021. PMID: 33927896 Free PMC article.
References
-
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42: 419–430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

