Global diversity of microbial communities in marine sediment
- PMID: 33077589
- PMCID: PMC7959581
- DOI: 10.1073/pnas.1919139117
Global diversity of microbial communities in marine sediment
Abstract
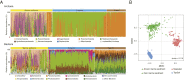
Microbial life in marine sediment contributes substantially to global biomass and is a crucial component of the Earth system. Subseafloor sediment includes both aerobic and anaerobic microbial ecosystems, which persist on very low fluxes of bioavailable energy over geologic time. However, the taxonomic diversity of the marine sedimentary microbial biome and the spatial distribution of that diversity have been poorly constrained on a global scale. We investigated 299 globally distributed sediment core samples from 40 different sites at depths of 0.1 to 678 m below the seafloor. We obtained ∼47 million 16S ribosomal RNA (rRNA) gene sequences using consistent clean subsampling and experimental procedures, which enabled accurate and unbiased comparison of all samples. Statistical analysis reveals significant correlations between taxonomic composition, sedimentary organic carbon concentration, and presence or absence of dissolved oxygen. Extrapolation with two fitted species-area relationship models indicates taxonomic richness in marine sediment to be 7.85 × 103 to 6.10 × 105 and 3.28 × 104 to 2.46 × 106 amplicon sequence variants for Archaea and Bacteria, respectively. This richness is comparable to the richness in topsoil and the richness in seawater, indicating that Bacteria are more diverse than Archaea in Earth's global biosphere.
Keywords: marine sediment; microbial diversity; subseafloor life.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Parkes R. J.et al., A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar. Geol. 352, 409–425 (2014).
-
- Parkes R. J.et al., Deep bacterial biosphere in Pacific Ocean sediments. Nature 371, 410–413 (1994).
-
- D’Hondt S.et al., Presence of oxygen and aerobic communities from seafloor to basement in deep-sea sediment. Nat. Geosci. 8, 299–304 (2015).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

