Bacteriophage SP01 Gene Product 56 Inhibits Bacillus subtilis Cell Division by Interacting with FtsL and Disrupting Pbp2B and FtsW Recruitment
- PMID: 33077634
- PMCID: PMC7950406
- DOI: 10.1128/JB.00463-20
Bacteriophage SP01 Gene Product 56 Inhibits Bacillus subtilis Cell Division by Interacting with FtsL and Disrupting Pbp2B and FtsW Recruitment
Abstract
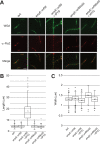
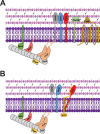
Previous work identified gene product 56 (gp56), encoded by the lytic bacteriophage SP01, as being responsible for inhibition of Bacillus subtilis cell division during its infection. Assembly of the essential tubulin-like protein FtsZ into a ring-shaped structure at the nascent site of cytokinesis determines the timing and position of division in most bacteria. This FtsZ ring serves as a scaffold for recruitment of other proteins into a mature division-competent structure permitting membrane constriction and septal cell wall synthesis. Here, we show that expression of the predicted 9.3-kDa gp56 of SP01 inhibits later stages of B. subtilis cell division without altering FtsZ ring assembly. Green fluorescent protein-tagged gp56 localizes to the membrane at the site of division. While its localization does not interfere with recruitment of early division proteins, gp56 interferes with the recruitment of late division proteins, including Pbp2b and FtsW. Imaging of cells with specific division components deleted or depleted and two-hybrid analyses suggest that gp56 localization and activity depend on its interaction with FtsL. Together, these data support a model in which gp56 interacts with a central part of the division machinery to disrupt late recruitment of the division proteins involved in septal cell wall synthesis.IMPORTANCE Studies over the past decades have identified bacteriophage-encoded factors that interfere with host cell shape or cytokinesis during viral infection. The phage factors causing cell filamentation that have been investigated to date all act by targeting FtsZ, the conserved prokaryotic tubulin homolog that composes the cytokinetic ring in most bacteria and some groups of archaea. However, the mechanisms of several phage factors that inhibit cytokinesis, including gp56 of bacteriophage SP01 of Bacillus subtilis, remain unexplored. Here, we show that, unlike other published examples of phage inhibition of cytokinesis, gp56 blocks B. subtilis cell division without targeting FtsZ. Rather, it utilizes the assembled FtsZ cytokinetic ring to localize to the division machinery and to block recruitment of proteins needed for septal cell wall synthesis.
Keywords: Bacillus subtilis; FtsZ; SP01; cell division.
Copyright © 2020 American Society for Microbiology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

