Airspace Macrophages and Monocytes Exist in Transcriptionally Distinct Subsets in Healthy Adults
- PMID: 33079572
- PMCID: PMC8048748
- DOI: 10.1164/rccm.202005-1989OC
Airspace Macrophages and Monocytes Exist in Transcriptionally Distinct Subsets in Healthy Adults
Abstract
Rationale: Macrophages are the most abundant immune cell in the alveoli and small airways and are traditionally viewed as a homogeneous population during health. Whether distinct subsets of airspace macrophages are present in healthy humans is unknown. Single-cell RNA sequencing allows for examination of transcriptional heterogeneity between cells and between individuals. Understanding the conserved repertoire of airspace macrophages during health is essential to understanding cellular programing during disease.Objectives: We sought to determine the transcriptional heterogeneity of human cells obtained from BAL of healthy adults.Methods: Ten subjects underwent bronchoscopy with BAL. Cells from lavage were subjected to single-cell RNA sequencing. Unique cell populations and putative functions were identified. Transcriptional profiles were compared across individuals.Measurements and Main Results: We identify two novel subgroups of resident airspace macrophages-defined by proinflammatory and metallothionein gene expression profiles. We define subsets of monocyte-like cells and compare them with peripheral blood mononuclear cells. Finally, we compare global macrophage and monocyte programing between males and females.Conclusions: Healthy human airspaces contain multiple populations of myeloid cells that are highly conserved between individuals and between sexes. Resident macrophages make up the largest population and include novel subsets defined by inflammatory and metal-binding profiles. Monocyte-like cells within the airspaces are transcriptionally aligned with circulating blood cells and include a rare population defined by expression of cell-matrix interaction genes. This study is the first to delineate the conserved heterogeneity of airspace immune cells during health and identifies two previously unrecognized macrophage subsets.
Keywords: RNA sequencing; alveolar macrophages; bronchoalveolar lavage.
Figures
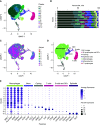
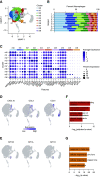
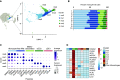
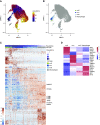

Comment in
-
What Is "Normal" When Examining Myeloid Cells in Human Airways?Am J Respir Crit Care Med. 2021 Apr 15;203(8):931-932. doi: 10.1164/rccm.202010-3932ED. Am J Respir Crit Care Med. 2021. PMID: 33181037 Free PMC article. No abstract available.
References
-
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;173:1307. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

