Rapid prediction of crucial hotspot interactions for icosahedral viral capsid self-assembly by energy landscape atlasing validated by mutagenesis
- PMID: 33079933
- PMCID: PMC7598928
- DOI: 10.1371/journal.pcbi.1008357
Rapid prediction of crucial hotspot interactions for icosahedral viral capsid self-assembly by energy landscape atlasing validated by mutagenesis
Abstract
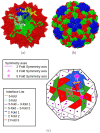
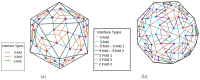
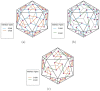
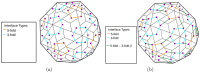
Icosahedral viruses are under a micrometer in diameter, their infectious genome encapsulated by a shell assembled by a multiscale process, starting from an integer multiple of 60 viral capsid or coat protein (VP) monomers. We predict and validate inter-atomic hotspot interactions between VP monomers that are important for the assembly of 3 types of icosahedral viral capsids: Adeno Associated Virus serotype 2 (AAV2) and Minute Virus of Mice (MVM), both T = 1 single stranded DNA viruses, and Bromo Mosaic Virus (BMV), a T = 3 single stranded RNA virus. Experimental validation is by in-vitro, site-directed mutagenesis data found in literature. We combine ab-initio predictions at two scales: at the interface-scale, we predict the importance (cruciality) of an interaction for successful subassembly across each interface between symmetry-related VP monomers; and at the capsid-scale, we predict the cruciality of an interface for successful capsid assembly. At the interface-scale, we measure cruciality by changes in the capsid free-energy landscape partition function when an interaction is removed. The partition function computation uses atlases of interface subassembly landscapes, rapidly generated by a novel geometric method and curated opensource software EASAL (efficient atlasing and search of assembly landscapes). At the capsid-scale, cruciality of an interface for successful assembly of the capsid is based on combinatorial entropy. Our study goes all the way from resource-light, multiscale computational predictions of crucial hotspot inter-atomic interactions to validation using data on site-directed mutagenesis' effect on capsid assembly. By reliably and rapidly narrowing down target interactions, (no more than 1.5 hours per interface on a laptop with Intel Core i5-2500K @ 3.2 Ghz CPU and 8GB of RAM) our predictions can inform and reduce time-consuming in-vitro and in-vivo experiments, or more computationally intensive in-silico analyses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Caspar DL, Klug A. Physical principles in the construction of regular viruses In: Cold Spring Harbor symposia on quantitative biology. vol. 27 Cold Spring Harbor Laboratory Press; 1962. p. 1–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

